Numerous molecular cloning techniques are currently available that allow for the insertion of a specific DNA fragment, or in some cases more than one fragment, into a vector of choice. Some of these cloning methods employ a restriction enzyme, which recognizes specific base pair sequence and cuts both DNA strands on both the vector and insert to create compatible ends, allowing the two to be joined together by DNA ligase. One of these methods is Golden Gate cloning.
What is Golden Gate cloning?
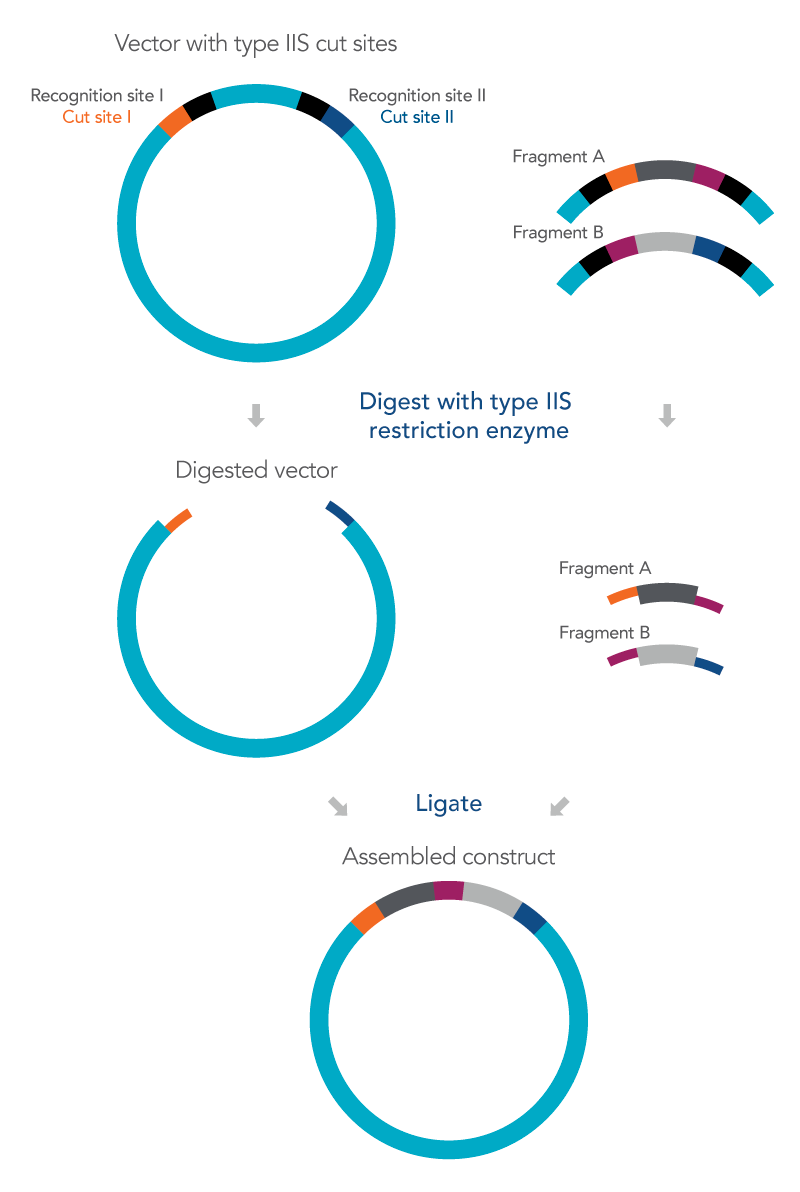
Golden Gate cloning utilizes Type IIS restriction enzymes to cut the insert and vector outside of their recognition site allowing for several fragments to then be assembled together with the vector in one reaction while at the same time also avoiding the formation of a scar in the final construct [1]. The overhangs produced using a Type IIS restriction enzyme are independent of the recognition site and unique to the sequence of the DNA fragments so after ligation no residual bases of the recognition sequence will be present in the cloned construct, preventing a so-called recognition site scar [1].
Because different Type IIS restriction enzymes cut the DNA at different locations outside of their recognition site, overhang sequences of differing lengths can be produced. By carefully designing the sequences based on the IIS restriction enzymes to be used, multiple adjacent fragments can be assembled together into a vector in a single ligation reaction [2]. To accomplish this, it is important to design DNA fragments with Type IIS recognition sites at the terminal ends that overlap with the adjacent fragments.
Applications of Golden Gate cloning
Golden Gate cloning is scarless and can be used for cloning single inserts, assembling large constructs originating from multiple fragments, as well as assembling fragments that contain internal structural complexities [1]. It is considered an efficient method that can be done in a single tube and, as it works better than some other methods with a wide range of fragment lengths, it lends itself to different levels of assembly. For example, it can be used to assemble several short genetic elements together to create a single gene, such as promoters, open reading frames, and terminators, or can be used to rapidly construct multiple genes into a vector simultaneously. It can even be used in to generate multigene constructs from a library of assembled genes [3].
Limitations
Areas that will not work well for Golden Gate cloning include sequences that contain GC extremities or secondary structures in the overlap regions. Although it can work better with repeat sequences than other scarless cloning methods that rely on homology alone, for Golden Gate assembly to work, it is critical that the Type IIS enzyme recognition site is uniquely present in the cloning site and does not appear anywhere else in the DNA sequence. If the recognition site occurs anywhere else in the insert or the vector, it will need to be removed to prevent undesired cuts. If there is an overhang with similar sequences (e.g., only 1 base pair difference) there is the chance that the vector may re-ligate which will decrease the cloning efficiency.
Design Considerations
- When designing DNA fragment(s) for Golden Gate cloning, include the cut sequence of the Type IIS enzyme in the terminal regions of the insert. Ideally, sequences containing 4-base overhangs have the highest accuracy. Add the restriction enzyme recognition site terminal to the cut site. Make sure the recognition and cut sites are also added to the vector so that the overlap joins together.
- Some enzymes require a spacer sequence between the recognition site and the cut site. For restriction enzymes that cut the sense strand one base pair outside of the recognition site, include a one base pair spacer nucleotide after the recognition sequence and before the cut sequence. For enzymes that cut after two base pairs, 2 base pair spacer nucleotides need to be included, and so on.
- Also, a 4–5 base pair flanking sequence facilitates the recruitment of enzymes to the DNA template. Hence, add such a sequence to both terminal ends of the fragment to increase reaction efficiency.
- Alternatively, plasmids can be used as starting material for Golden Gate cloning reactions. IDT can provide requested sequences in cloning vectors that are suitable for Golden Gate cloning (i.e. no Type IIS restriction sites present outside of the cut site).


