What is colony PCR?
Colony PCR is a method to quickly check bacterial colonies generated after the transformation step of a molecular cloning protocol [1,2]. It is used to verify that the vector inserted during transformation contains the full desired genetic insert. Other methods used to screen clones may require plasmid DNA extraction and some level of purification, followed by a digestion reaction with a restriction enzyme, and verification of a specific DNA banding pattern on an agarose or acrylamide gel. Because colony PCR uses the plasmids present in the lysed cells as the DNA template for the PCR reaction these extra steps are unnecessary.
How is colony PCR done?
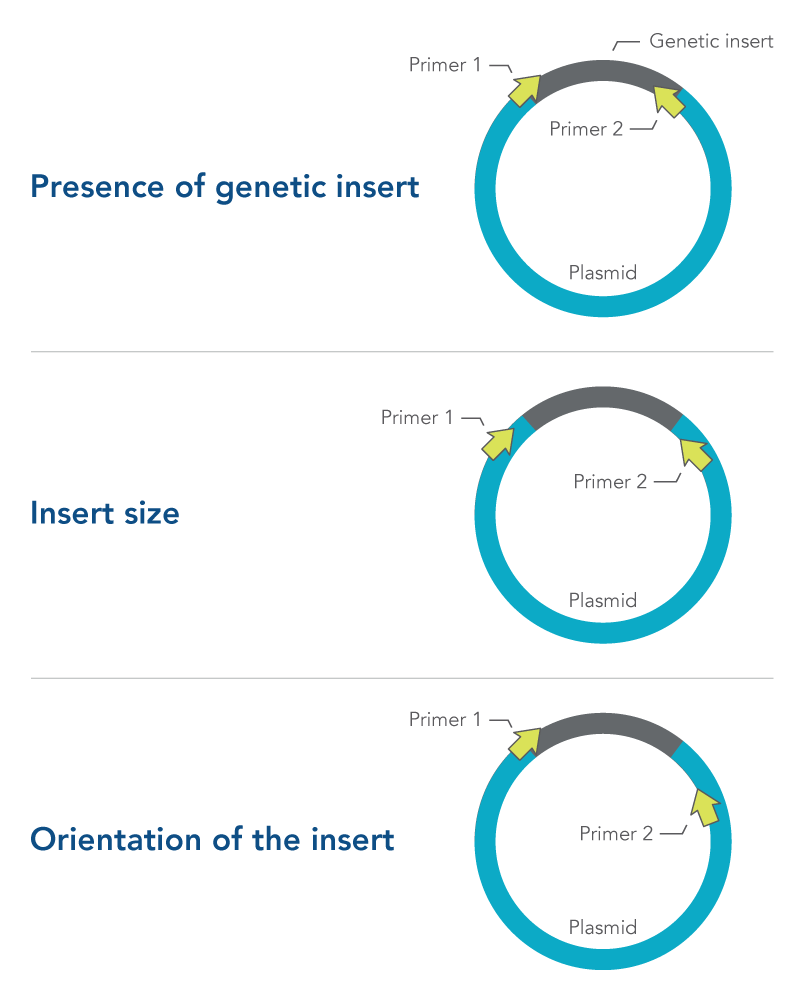
Primer design is especially important in colony PCR. Depending on what they are designed to target, primers selected for colony PCR can provide information about:
- Presence of genetic insert—primers are designed to target the insert and not the plasmid
- Insert size—primers are designed to flank the insert
- Orientation of the insert—one primer is designed to target the insert, while the other primer targets the plasmid
All three of these approaches require careful primer design and testing (Figure 1). More information about primer design can be found in the DECODED article, How to design primers and probes for PCR and qPCR.
After transformation, the colonies are picked with a sterile pipette tip, toothpick, or wooden applicator stick. Generate either a grided replicate plate or a 96 deep well plate with growth media. Either of these can act as a reference source for useful colonies after running the PCR reaction. The remaining cells on the pipette tip or wooden applicator stick can serve as the DNA template for a colony PCR reaction. Based on the number of colonies screened, make up enough bulk reaction mix and aliquot into PCR tubes or 96 well plate. Swirl the pipette tip or wooden applicator into a well to add the cells. During the thermocycling the cells will break open and release the plasmid template.
After thermocycling the PCR product is visualized using an agarose gel, or capillary based DNA analyzer. The colony PCR product should be of the expected size and should represent a single clear band on the gel. If multiple bands are present or if the band is not in the correct size range, the colony tested likely does not contain the correct insert in the plasmid.
If the band is in the correct size range on the gel and only one band is present, it is likely that the plasmid contains the desired insert. The final verification step is to sequence this PCR product to ensure that no other small changes occurred in the insert (e.g., SNPs).
Tips for colony PCR
Colony PCR offers researchers a quick, straightforward method for screening clones generated during the molecular cloning process.
Here are 3 tips for your colony PCR:
- Avoid using too much starting material. It’s important when adding the lysed cells to the PCR reaction mix that too much material isn’t added as the DNA template as this can result in nonspecific amplification.
- Include controls. Both positive and negative controls should be included in colony PCR. A good positive control would be any DNA that you are sure should amplify using the primers selected, this helps to ensure that the PCR reaction ‘worked’, i.e., that the reagents used haven’t expired, etc. It is also recommended that a negative control is included – one reaction that hasn’t been exposed to a bacterial colony to ensure that your PCR reagents are clean.
- Beware of false positives. It’s a good idea to test multiple colonies and sequence all ‘positive’ results. Colony PCR can provide general information about the size of the amplified product; however, sequencing is important to ensure that small genetic edits such as SNPs have not been incorporated into your insert during the cloning process.