Recent advances in genome engineering technologies based on the CRISPR-Cas9 System enable opportunities for systemic interrogation of gene functions in mammalian cells. CRISPR-Cas9 genome editing tools can be used to introduce a change at a specific genomic location, either to cause a frameshift for loss-of-function studies or to insert an exogenous sequence for gain-of-function studies.
Similar to other nuclease-driven mutagenesis methods that introduce site-specific nucleotide changes, CRISPR-Cas9 editing in tissue culture results in a heterogeneous, polyclonal population in which the editing outcome may differ among individual cells. In some experimental settings, it may be necessary to generate an edited cell line containing a homogeneous genetic background to retain the desired genotype, and to facilitate downstream phenotypic characterization.
Isolating individual clones from a pooled population
Multiple approaches have been developed for isolation of monoclonal cell lines, such as single-cell sorting, isolation with cloning cylinders, and limiting dilution [1–3]. Although these methods are well-established and widely used, they all have limitations. Fluorescence-activated cell sorting (FACS) requires a fluorescent reporter to be co-delivered and expressed, while cloning cylinders are used exclusively for adherent cells.
Limiting dilution requires a highly diluted cell suspension from which single cell-derived clones are isolated and further expanded. Unlike single-cell sorting or using cloning cylinders, which require sophisticated instruments, limiting dilution can be done with standard pipetting tools. However, it is a laborious and time-consuming process with low throughput. Also, this method cannot be applied to cells that do not propagate from single-cell clones. The probability of obtaining a single-cell in an aliquot of statistical value makes this method inherently inefficient. So, while limiting dilution provides the most versatile approach, it will require adjustments to give you the best chance of successful single clone isolation.
Comparison of 3 limiting dilution cloning protocols
IDT scientists assessed three types of limiting-dilution cloning protocols: low-density seeding, serial dilution, and array dilution. Of those, the array dilution method provides the highest success rate combined with ease of use [4]. This method can be used to efficiently isolate a monoclonal cell line from a recently CRISPR-Cas9 engineered cell population, followed by clonal expansion to generate the desired cell line. We provide a detailed, step-informed protocol focused on adherent cells in the Appendix below.
Low-density seeding method
The first limiting-dilution cloning method was performed using a diluted cell density as low as 0.5 cells per aliquot. This requires transferring 100 µL aliquots of a transfected cell suspension (5 cells/mL in complete growth medium) into each well of a 96-well plate. According to a Poisson distribution, seeding at an average of 0.5 cells/well ensures that at least some wells receive a single-cell, while minimizing the likelihood that any well receives more than 1 cell. This method was widely adopted for clonal isolation of hybridomas, but its efficiency in our hands was unexpectedly low, possibly due to relatively low cell viability after transfection.
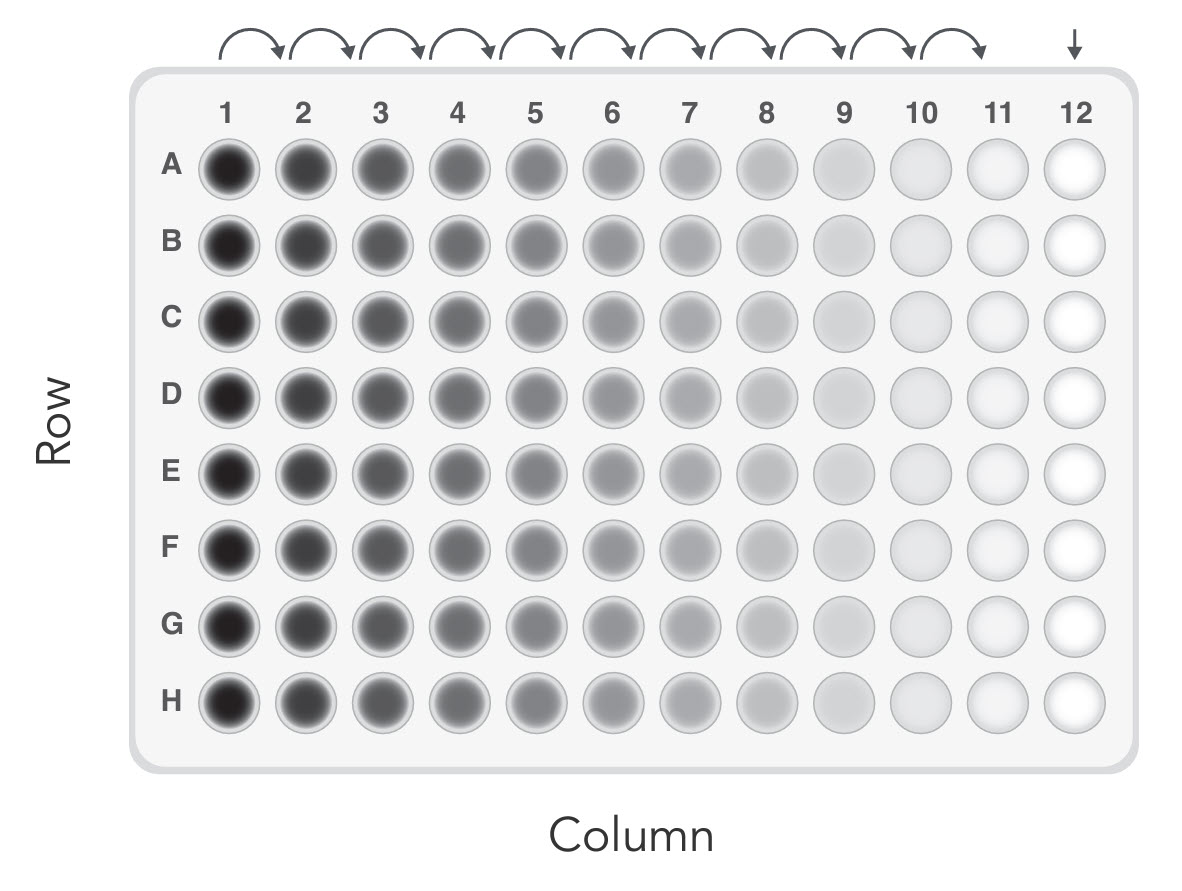
Serial dilution method
The second method we tested started with a much higher cell density. As shown in Figure 1, the wells in Column 1 received 100 µL of cell suspension at a density of 1000 cells/mL. Subsequently, 2-fold serial dilutions were made horizontally across the plate. Serial dilution avoids the need to pipette very small volumes so is commonly used to prepare diluted analytes. The wells receiving the initial inoculum can be used to help focus the microscope when scanning the plate for individual cells that are sometimes difficult to find. Single clones are expected in Column 11 and adjacent columns. Compared to the low-density seeding method, in which the single clones are randomly distributed across the plate, this method saves the scanning labor by limiting the possible wells of interest to 3–4 columns instead of the entire 96-well plate. We also observed a discernible improvement in efficiency with this serial dilution method compared to low-density seeding.
Array dilution method
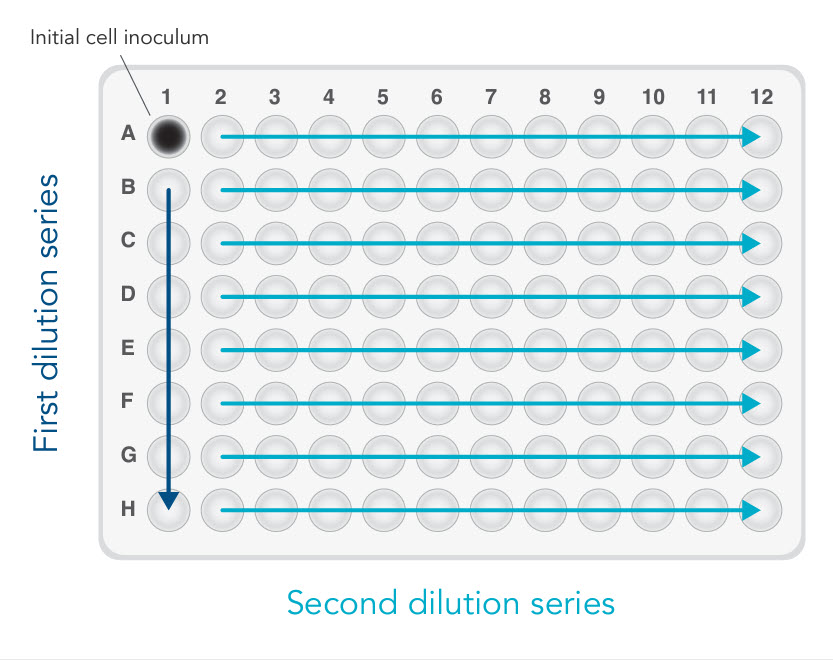
The last method, array dilution, provided us with the highest success rate [4]. The initial inoculation occurs in one well instead of an entire column (see Figure 2). This allows for a subsequent 2-fold serial dilution—first vertically, then horizontally. In our hands, we retrieved more than 30 single clones out of 3 x 96-well plates. As expected, most of the single clones were found along the diagonal.
See the detailed, step-informed array dilution protocol focused on adherent cells in the Appendix below.
Appendix: An array dilution procedure for isolating CRISPR-Cas9 edited single clones derived from HEK-293 cells
Important considerations before you start:
- Editing efficiency is affected by many experimental factors, such as cell type, guide RNA design, and delivery efficiency. It is important to first optimize your genome editing experiments in your cells. Perform a primary validation experiment to determine the relative fraction of cells containing an edit. Based on the editing efficiency, you can estimate the number of single clones you will need to screen to identify a clonal cell line carrying the desired mutation.
- The validation process before single clone isolation varies depending on the nature of the desired mutation. Small indels are often detected in a mismatch cleavage assay (using, for instance, the Alt-R™ Genome Editing Detection Kit). A single nucleotide change can be assayed by Sanger sequencing, next generation sequencing, or droplet digital PCR genotyping. Large insertions and deletions can be identified by a size change in the PCR product produced by primers flanking the region edited.
- The survivability of single cell clones varies by cell type. Ensure that your cell line can produce colonies. Not every cell line is able to grow under the conditions described in the Appendix protocol. Some cells require contact with one another to grow and tend not to grow very well in sparse cultures due to a lack of secreted growth factors.
- The number of monoclonal cells obtained using this method is dependent on a number of experimental factors, such as the growth properties of the cell used. We typically obtain 10–15 single clones in each 96-well plate with the use of CRISPR-Cas9 edited HEK-293 cells.
Step 1. Prepare cells before transfection.
Note: HEK-293 cells are cultured in DMEM medium containing 10% fetal bovine serum (FBS). Cells are maintained at 37°C in a CO2 incubator at 37°C and passaged every 3 days. Ideally, cells should be at 70–80% confluency at the time of transfection.
Step 2. Deliver Alt-R ribonucleoprotein (RNP) complexes into HEK-293 cells in a 6-well plate.
A. (Recommended) Perform the transfection in a 6-well plate to ensure sufficient cells are available for subsequent experiments.
B. Incubate the transfected cells at 37°C in a 5% CO2 incubator for 48 hours, or until cells are 70–80% confluent.
Notes:
Lipofection is a method to deliver CRISPR-Cas9 RNP to HEK-293 cells. See the user guide, Alt-R CRISPR-Cas9 System: Cationic lipid delivery of CRISPR ribonucleoprotein complex into mammalian cells for the recommended experimental setup and detailed protocol.
Electroporation may be required for some cell types that are refractory to lipid-mediated transfection, or are susceptible to cytotoxicity from the lipid reagents. Refer to the user guide Alt-R CRISPR-Cas9 System: Delivery of ribonucleoprotein complexes into HEK-293 cells using the Amaxa® Nucleofector® System or the user guide Alt-R CRISPR-Cas9 System: Delivery of ribonucleoprotein complexes into Jurkat T cells using the Neon® Transfection System for the recommended experimental setup and detailed protocol.
Step 3. Resuspend transfected cells to an optimal density.
A. Aspirate the medium and wash cells twice with pre-warmed PBS.
B. Add 500 µL of 0.05% Trypsin-EDTA to each well of the 6-well plate. Incubate the plate at 37°C in a CO2 incubator for 2–5 minutes. Use a microscope to verify that cells detach from the plate.
C. Stop trypsinization by adding 1 mL DMEM medium supplemented with 10% FBS. Break up any cell clumps into individual cells by passing cells several times through a serological pipet. Transfer the cell suspension to a sterile 15 mL centrifuge tube.
D. Count cells in the suspension to determine cell density. Dilute the suspension for more accurate counting as necessary.
E. Determine the total number of cells required for your experiment. Further dilute cells in complete growth medium to 2 x 104 cells/mL or the concentration suited to your cells.
Tip: We recommend 2 x 104 cells/mL as a good starting point. Further titration may be required for other cell types.
Step 4. Isolate single clones via serial dilution.
A. As shown in Figure 2, add 100 µL of complete culture medium to all wells of a 96-well plate, except well A1 (Figure 2).
B. Add 200 µL of cell suspension with proper cell density to well A1.
Note: For an input concentration of 2 x 104 cells/mL, the total cell number is 4000.
C. Transfer 100 µL of cell suspension from well A1 to B1. Mix by gently pipetting. Repeat the 1:2 dilutions down Column 1 using the same pipet tip (Figure 2, first dilution series).
D. Add 100 µL of complete culture medium to wells A–G in Column 1 to reach a final volume of 200 µL/well. Mix by gently pipetting.
E. Use the same tips to transfer 100 µL of cell suspension across the plate horizontally from Column 1 to Column 2. Mix by gently pipetting. Repeat the 1:2 dilution across the rows using the same pipet tips (Figure 2, second dilution series).
F. Add 100 µL of complete culture media to all wells in Columns 1–11 to bring the final volume of each well to 200 µL.
G. Incubate the plate at 37°C in a CO2 incubator. After 4–5 days, check cell growth and mark wells containing just 1 single colony. Incubation time may vary depending on cell growth rate.
H. Monitor cell growth daily and transfer colonies into larger vessels.
Note: HEK-293 cells are typically ready for subculture within 2 weeks after seeding on the 96-well plate.
Tip: Subculture a portion of the cells derived from the single clone into a larger vessel (e.g., 24- or 48-well plate). Transfer the rest of the cells into a new 96-well plate for screening purposes. Depending on the nature of the desired nucleotide change, various methods such as Sanger sequencing, next generation sequencing, or genotyping via qPCR can be used for confirmation of the desired genome editing events.