The goal of a qPCR experiment is to amplify a target sequence using a set of primers. Using tools such as NCBI BLAST, PrimerQuest™ Tool, Predesigned qPCR Assays, and RealTime qPCR Assay Entry, you can quickly and easily design a successful qPCR experiment. Whether you are interested in alternatively spliced variants, copy number variations, or single nucleotide polymorphisms, you will need to understand how to design your primers correctly.
How to design primers for qPCR
To improve the success of your qPCR experiment it is necessary to design good primers that are complementary to the nucleotide sequence of interest. You will need two primers, one forward and one reverse, to amplify the target DNA.
You want to design exon-exon primers since they are specific for amplifying cDNA, not genomic DNA (gDNA) (Figure 1). The exons contain coding sequences while introns are non-coding sequences. By designing primers that target the 5’ end of one exon and the 3’ end of the neighboring exon, this will improve the efficiency for your qPCR experiments.
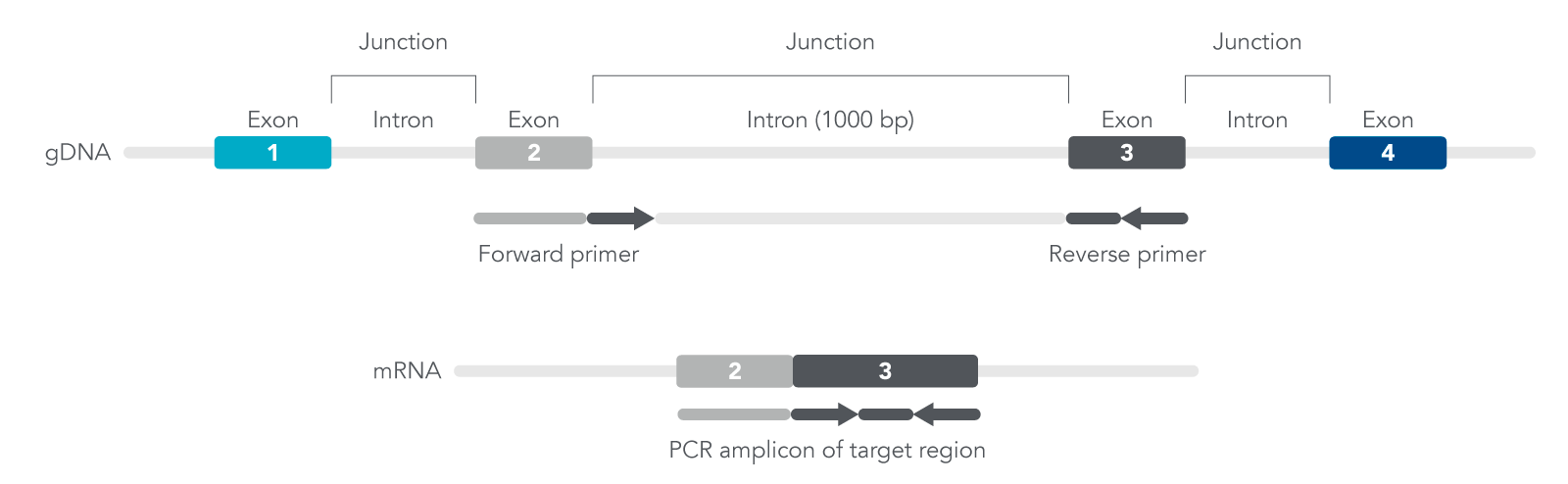
Figure 1. Overview of how qPCR exon-exon primers are designed based on location of exons and introns.
Here are 6 useful tips for how to design good qPCR primers that span an exon-exon junction:
Design primers for various isoforms of the target sequence
For your sequence, identify the location of the exons and design primers where one exon is the forward primer while the second exon is the reverse primer, or the second exon is the forward and third exon is the reverse primer, and so forth (Figure 1). Doing so may result in either longer or shorter isoforms derived from the same target region of interest. Use our PrimerQuest Tool to validate your primer pairs and quickly design your qPCR assay.
GC content
Having a high GC content of 40–60 % improves stability. The number of the Gs and Cs in the primer refers to a percentage of the total bases. It’s necessary to have a G or C residue at the 3’ end of the primer to improve the binding to the DNA template. When designing your primers, you will need to avoid having more than 3 G’s or C’s because this can lead to primer-dimer formation.
Primer length
A primer pair that is specifically designed to span an exon-exon junction anneals and amplifies at the correct target region within the transcript. The forward and reverse primers that will bind upstream and downstream of the exon should be between 18–24 bp in length. A longer sequence will decrease the efficiency of the experiment since a longer primer will take longer to hybridize, extend, and remove.
Melting temperature (Tm)
When designing primers, the ideal melting temperature is 60–64°C. This temperature is based on the cycling and reaction conditions. The Tm of your primer pair should not differ more than 2 °C to ensure that primers bind and amplify the PCR product. You can determine the Tm of your primers by using IDT’s OligoAnalyzer tool.
Annealing temperature (Ta)
The annealing temperature (Ta) of your primers should be 5°C lower than the Tm for your primers. If the Ta is too low for either primer or both primers this can result in primers annealing to sequences other than your target sequence leading to nonspecific PCR amplification. Therefore, to ensure optimal annealing temperature for your primers use our OligoAnalyzer tool.
Amplicon size
If the PCR amplicon is too large, this can result in gDNA contamination. The optimal amplicon size for qPCR is 75–150 bp.
Read our DECODED article on How to use the PrimerQuest tool to design your qPCR experiments quickly and easily. If you need additional assistance with designing primers for your qPCR experiment, contact us.
RUO24-2717_001

