Luciferases are a category of enzymes that emit light when oxidation occurs. It is an enzyme that is made in cells of specific systems to regulate bioluminescence. These enzymes are found in a variety of organisms such as fireflies, marine organisms, algae, fungi, and bacteria and are commonly used as reporters in biological experiments [1]. An assay using luciferase can be used to study gene expression as well as gene regulation and because it is extremely responsive, very small changes in transcription can be determined using this technique [2]. In addition, it is an alternative to using fluorescence to study these processes as it provides several advantages including decreased background interference or minimal autofluorescence as well as higher amplification and a dynamic range of signal produced from the assay [2,3].
One of the first luciferase enzymes to be investigated and subsequently used as a reporter construct was from the North American firefly Photinus pyralis. The oxidation reaction of this luciferase requires oxygen, ATP, and the luciferin substrate where the resulting excited state of oxyluciferin leads to bioluminescence [1]. For the firefly this is used for reproduction while in the lab this reaction is helpful for a wide variety of genetic applications. There are many other species that also use the luciferase enzyme; however, this enzyme is highly diverse and in each case the substrate and structure can vary so research is ongoing into many other organisms and their specific luciferase enzymes to better understand how to use them in research applications [1]. Table 1 includes some of the most common species used as Luciferase reporters along with the organism it is found in, size of the protein, substrates needed for the oxidation reaction to occur, and the name of the luciferase.
Table 1. Common Luciferase reporters.
| Species | Organism | Size(kDa) | Substrate | Luciferase |
|---|---|---|---|---|
| Cypridina noctiluca | Ostracod | 62 | Vargulin | Cluc |
| Gaussia princeps | Copepod | 20 | Coelenterazine | Gluc |
| Renilla reniformis | Sea pansy | 36 | Coelenterazine | Rluc |
| Photinus pyralis | Firefly | 61 | Luciferin | Fluc |
Luciferase gene and protein
As discussed above, a variety of organisms utilize luciferase gene regulation for an array of light generating reactions and thus the proteins encoded by the luciferase gene also differ based upon species. As shown in Table 1, the size of the proteins differs ranging from 20 to 62 kDa in just the four species shown. Additionally, the structures and folding patterns are also quite variable from one species to the next. For example, the luciferase from the firefly P. pyralis folds into two distinct domains forming a unique anvil and hammer motif not seen in other luciferase enzymes. In comparison, bacterial luciferase folds into a common and well-known TIM barrel structure [1].
Luciferase function
The overall function of luciferase is to regulate bioluminescence. In the lab for research purposes, it can be used as a light reporter for assays developed to study promoter activity, transcription, or enzyme activity. It can also be used for imaging studies or to detect ATP levels and can be used in cell viability assays as well as protein denaturation studies. It provides many useful experimental opportunities.
Luciferase assay
The luciferase assay is a useful tool that can be used to look at gene expression in the laboratory. One beneficial use is to look at whether activation or repression of a gene is taking place by a protein. This can be done using the luciferase assay because it makes an association between a protein being present and the resulting amount of gene product made [4]. Using the assay for this purpose requires making a construct that includes the promoter region of your gene of interest (the target gene) and fusing it with the coding sequence for luciferase. In addition, a second DNA construct is also needed that codes for the protein thought to be involved in regulating transcription of your target gene [4].
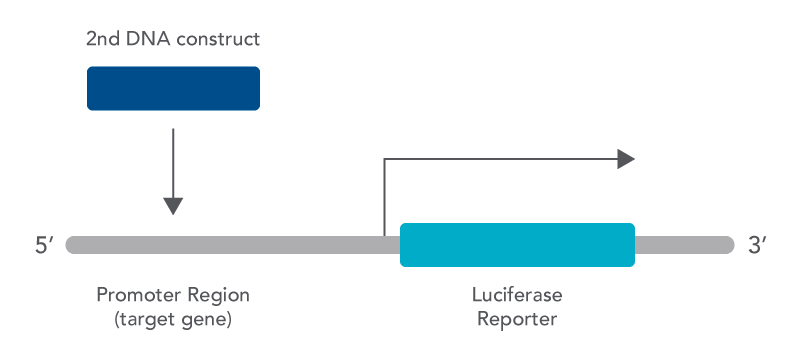
There are many other ways that the luciferase assay can also be used as this is just one example. In addition, luciferase from many different organisms can be selected for your assay depending on the nature of the experiment being conducted. Factors that need to be considered include the following:
- The substrate used by the luciferase for that organism.
- The flash kinetics (sensitivity) of that enzyme.
- The glow kinetics (stability) of your chosen enzyme.
- The wavelength at which the luciferase emits light.
- Whether or not the enzyme is dependent upon ATP for oxidation.
Selecting the assay that is optimal for your type of study is critical for optimal results. Imaging studies are better suited for luciferases that emit light at wavelengths >600 nm and cell viability studies are best for luciferases that are ATP-dependent [5].
GFP Luciferase
For some species, close coupling of luciferase with GFP is needed for bioluminescence to occur. This is the case with Renilla reniformis which is a bit more complex that some of the other luciferase reactions. This particular luciferase has a GFP protein that interacts closely to fully complete a visible reaction. Initially when the coelenterazine substrate is oxidized, the blue light that is emitted is not visible in vivo. It requires another reaction which results in green bioluminescence that is now visible in the animal [6].
How can IDT help?
IDT offers a variety of products and tools that can help you get started generating your constructs for your luciferase assays. Depending on the size of the construct you are looking for, IDT provides gene fragment products that may meet your needs. Check out our gBlocks™ and eBlocks™ Gene Fragments to see if one of these products might help with your experiments. If you want to order your construct as a plasmid IDT offers that option as well as a Gene and MiniGene™ product. If you are not sure what product is best for your work contact us.
Instead of spending days designing and constructing your synthetic gene, you can use the Codon Optimization Tool to order your gene in minutes through IDT and spend the saved time advancing your research while we make your genes or IDT Gene Fragments. Access IDT’s free online Codon Optimization Tool and get started. We also provide a step-by-step video tutorial for using the Codon Optimization Tool.
Contact us
If you are working with an organism not listed in the Codon Optimization Tool’s Organism list, or do not see the information you need, contact us. We can accept non-standard optimizations that fall outside of the rules used by the tool (design fee may apply).

