The discovery of the DNA structure
The milestone discovery of the double helix structure of deoxyribonucleic acid (DNA) gave rise to the field of molecular biology. In 1953, James Watson and Francis Crick, with the help of Rosalind Franklin’ s X-ray crystallography image of DNA, discovered DNA’s classical double helical structure [1]. Erwin Chargaff discovered four types of bases that make up DNA, which are purine bases adenine (A) and guanine (G), and the pyrimidine bases cytosine (C) and thymine (T) [1]. Chargaff’s rules are that A always pairs with T and C pairs with G.
Other discoveries made by Oswald Avery determined that DNA carried hereditary information in pneumococcal bacteria while Alexander Todd determined that the DNA molecules contain repeat phosphate and deoxyribose sugar groups [1].
Structure of DNA and RNA
In DNA or RNA, phosphodiester bonds are the bonds between the phosphate group and the sugar molecules. DNA contains nitrogenous bases (adenine, guanine, cytosine, thymine, or an uracil if it’s RNA), a sugar molecule (deoxyribose if DNA or ribose if RNA), and a triphosphate group (Figure 1).
Ribose is the sugar molecule in RNA and is a 5-carbon molecule (C5H10O5). In DNA, the deoxyribose sugar molecule contains one less oxygen atom (C5H10O4). The phosphate group attaches to the 5th carbon atom of the sugar molecule.
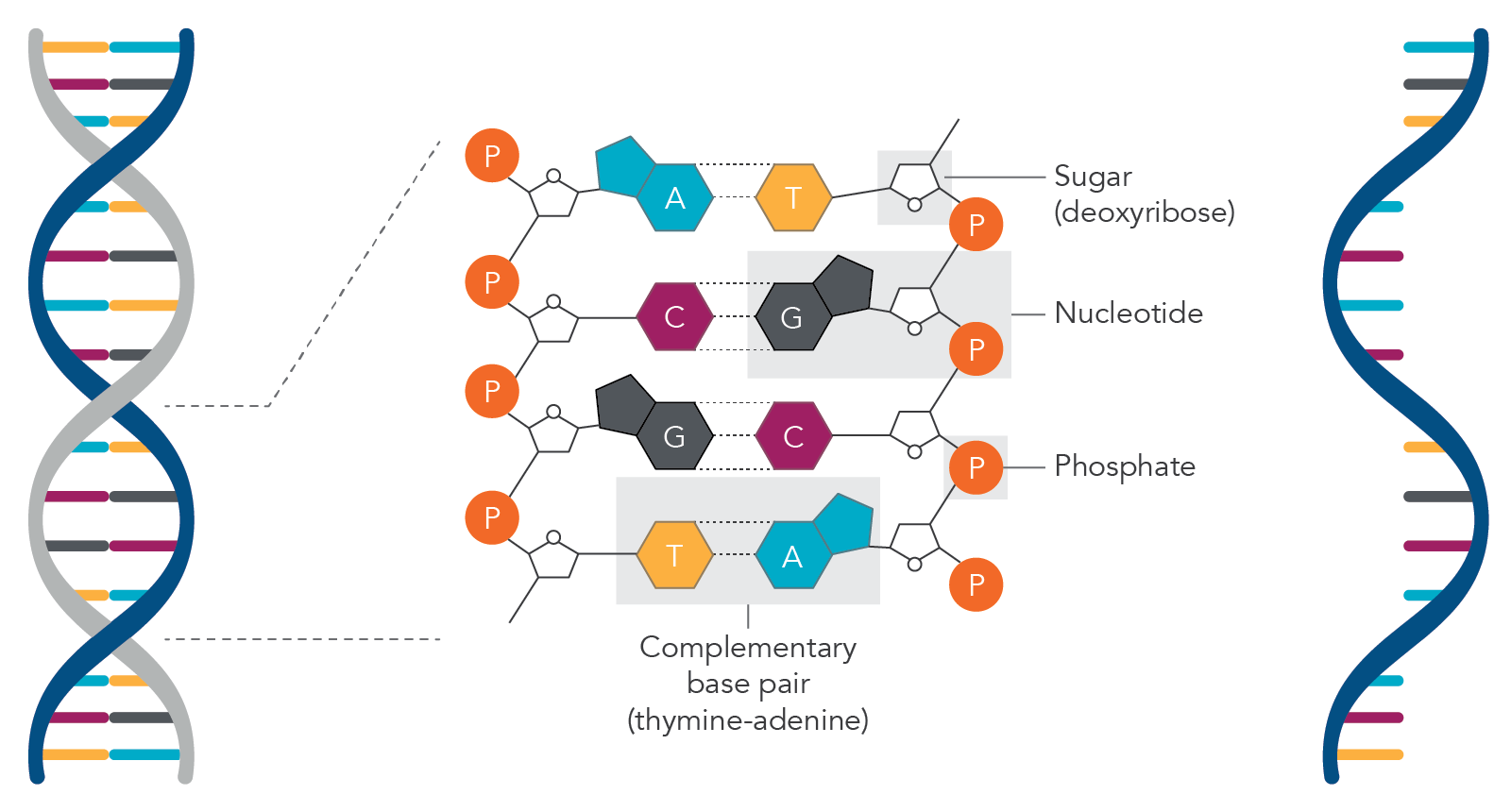
DNA (double-stranded) and RNA (single-stranded) molecules. Based on Chargaff rules for base paring in DNA, adenine (A) base pairs with thymine (T) and guanine (G) base pairs with cytosine (C). For RNA molecules, adenine (A) base pairs with uracil (U). In DNA, the sugar molecule is deoxyribose while in RNA the sugar molecule is ribose. When the base is attached to a sugar molecule it is known as a nucleoside and when the nucleoside is attached to the phosphate group, it is called a nucleotide.
Phosphodiester bonds
When the nitrogenous base is attached to a sugar molecule it is referred to as a nucleoside, and when the nucleoside is attached to the phosphate group it is called a nucleotide. The phosphate group is derived from phosphoric acid (H3PO4), and when attached, this will lead to the loss of two hydrogen atoms.
The bond formation occurs at the 5th carbon of the sugar molecule, which contains two hydrogen atoms and a hydroxyl group (-OH group) while the phosphate group loses a hydrogen (H) atom. When the bond is formed between them, a water molecule (H2O) is released, thus forming an ester bond. The phosphodiester bond is formed when the phosphoric acid molecule forms two ester bonds (Figure 2). The nucleotide molecule forms an ester bond when the nucleoside attaches to the phosphate group and continues to attach to neighboring sugar molecule attached to the nucleotide—this sugar-phosphate-sugar bond makes up the backbone of the DNA or RNA. The bond formation between two different nucleotides is caused by the loss of the hydroxyl group (-OH) from the 3rd carbon of the ribose sugar and the hydrogen (H) from the phosphoric acid, thus creating a second ester bond, which refers a phosphodiester bond.
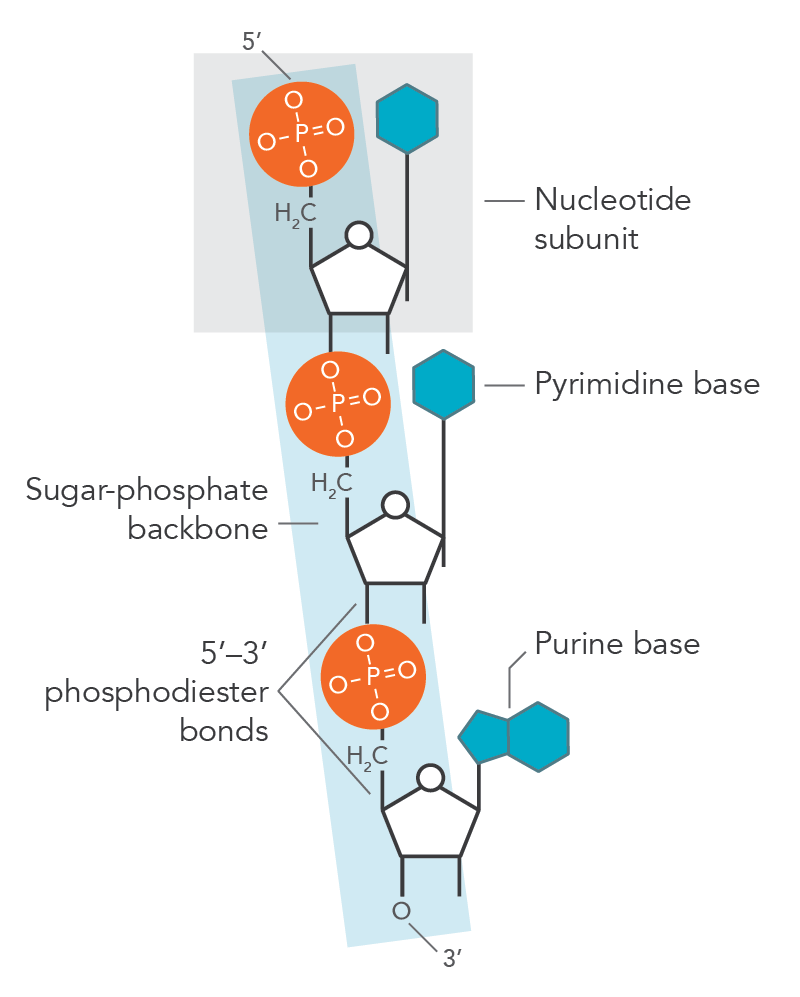
Phosphodiester bond. This is the sugar-phosphate-sugar bond that creates the backbone of the DNA and RNA molecules.
Learn more about how IDT’s oligos modifications can block nuclease degradation and phosphorothioate bond modifications.

