rhAmpSeq™ Index Primers
Combine amplicon libraries in a single sequencing run for maximum efficiency
Use rhAmpSeq Index Primers to prepare amplicon libraries for targeted sequencing on Illumina® platforms by adding a unique, identifying “barcode” sequence to each amplicon. Request exactly the primers you need from the 96 sample indexes available for P5 and P7 Illumina® index primer sequences. IDT scientists have tested and validated these 96 sample index sequences to ensure optimal performance.
Ordering
rhAmpSeq Index Primers
Add unique "barcode" sequences and P5/P7 sequences to your amplicon libraries. Priced per primer.
To complete your rhAmpSeq CRISPR workflow, you will also need a custom rhAmpSeq CRISPR panel and the rhAmpSeq CRISPR Library Kit. The rhAmpSeq CRISPR Library Kit includes Analysis Credits to enable data processing and quantification of editing events with the rhAmpSeq CRISPR Analysis Tool.
Product details
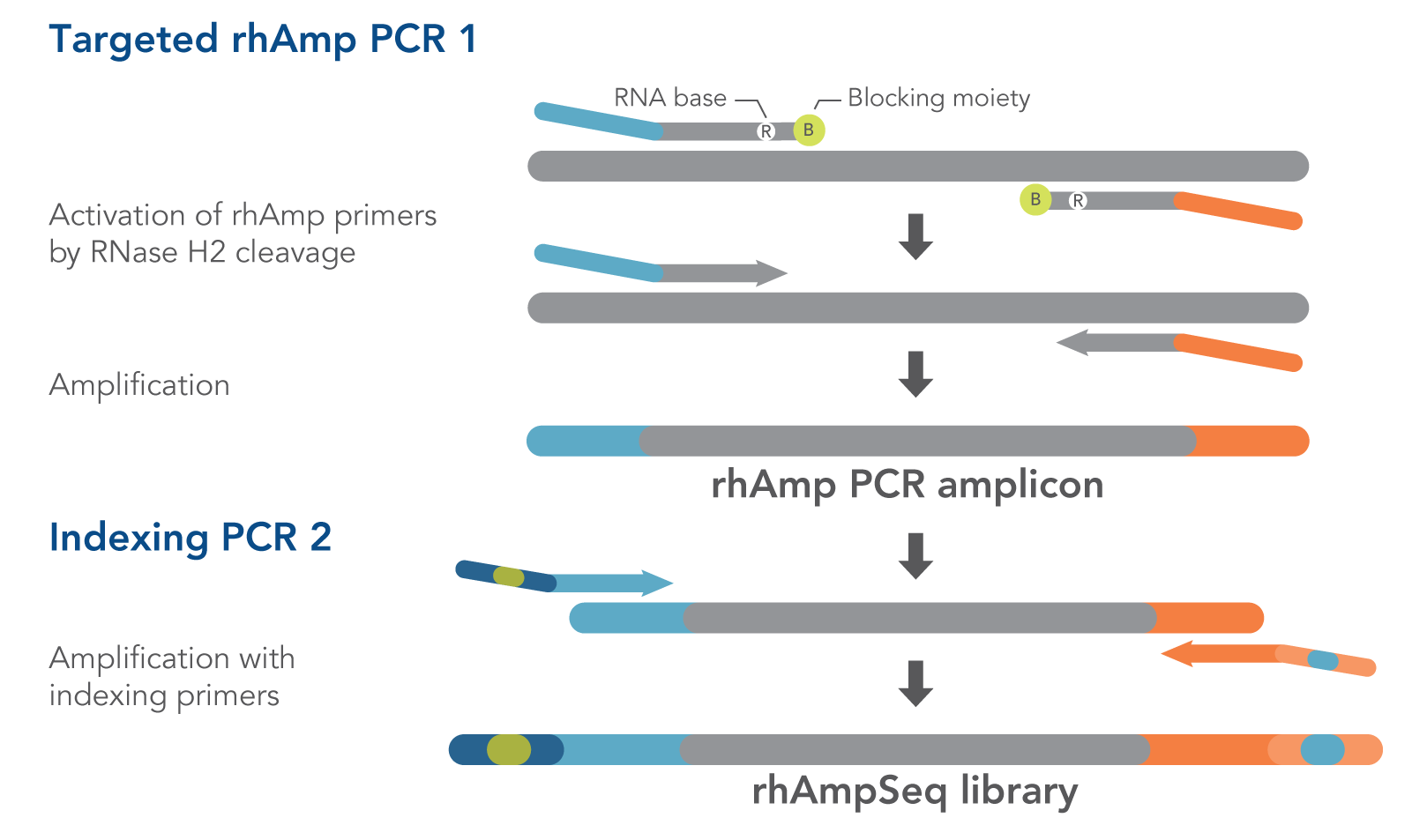
rhAmpSeq Index Primers are used in the second amplification step of the rhAmpSeq workflow, Indexing PCR 2, to add both unique index sequences and the P5/P7 sequences recognized by Illumina® sequencing instruments (Figure 1). These 96 index sequences are available for both the P5 and P7 primers. Adding these sequences to the amplicons created in the first amplification step, Targeted rhAmp PCR 1, creates dual-indexed libraries.
IDT scientists designed and tested the 96 index sequences for optimum performance.
Figure 1. Detail of amplification steps in the rhAmpSeq workflow. RNase H2 activates rhAmp primers by target-specific cleavage of the RNA base within the DNA:RNA duplex, removing a 3′ blocker. RNase H2 activity is highly specific, thus reducing the amount of amplification from non-specific hybridization and primer dimers. Only activated rhAmp primers can be extended to generate target amplicons.
Illumina® sample barcodes and P5/P7 sequences are incorporated during Indexing PCR 2.
System features and specifications
| Feature | Specification |
|---|---|
| Supported applications/protocols | On- and off-target analysis of genome editing experiments High-throughput analysis of on-target genome editing experiments |
| Insert size | Flexible (50–200 nt) |
| Custom panel size | Up to 5000 amplicons per panel |
| Sample indexing capability | 96 index sequences (up to 9216 combinations) |
| Compatible platforms | Illumina® |
| Hands-on time* | 1–1.5 hr |
| Data analysis time** | 0.5–2 hr |
* Estimated time to process 12–96 samples using manual pipetting, including reaction setup, cleanup, library quantification, and normalization steps
** Estimated time to process sequencing data (up to 500 amplicons) through rhAmpSeq CRISPR
Analysis Tool
Resources
Frequently asked questions
What sample types can I use with rhAmpSeq™ assays?
The rhAmpSeq assay is compatible with purified genomic DNA, cell-free DNA (cfDNA), and formalin-fixed, paraffin-embedded (FFPE) tissue.
What are the advantages of the rhAmpSeq™ Library Preparation Protocol and the rhAmpSeq High Throughput Library Preparation Protocol?
The rhAmpSeq™ Library Preparation protocol offers high assay performance (i.e., more mappable reads and better coverage uniformity between assays), while the rhAmpSeq™ CRISPR library preparation is a streamlined protocol that saves time and overall cost.
How much time does each rhAmpSeq™ CRISPR protocol require?
The rhAmpSeq CRISPR Library Preparation Protocol takes approximately 4–4.5 hours with 1–1.5 hours of hands-on time.
Note: Times listed are for processing 12–96 samples using manual pipetting, including reaction setup, cleanup, library quantification, and normalization steps. The time for either protocol will vary depending on the number of samples being processed.