xGen™ Pan-Cancer Hybridization Panel
Research the genes implicated in several cancers
The xGen Pan-Cancer Hyb Panel consists of 7,816 xGen Hyb Probes for research, spanning 800 kb of the human genome, for enrichment of 127 mutated genes implicated across 12 tumor tissue types for deeper sequencing coverage in research studies.
xGen NGS—made for cancer research.
Ordering
- Obtain high coverage uniformity across all targets
- Identify variations reliably with increased depth of coverage
- Enjoy fast turnaround (TAT) via easy online ordering and next-day shipping
Transform Your NGS Workflow with Automation
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
For the xGen Hybridization and Wash Kit, xGen Universal Blockers, xGen Library Amplification Primer Mix, and/or xGen Human Cot DNA, please visit the xGen Hybridization Capture Core Reagents page.
Product details
Every year, approximately 450 people in every 100,000 are diagnosed with cancer with a 5-year relative survival rate at 67.7%. Although survival rates have increased from less than 50% in 1975, there is a need for more basic research into the types of mutations and whether they are driver mutations or passenger mutations [1].
Research studies to produce a short list of mutated genes that are relevant to a multitude of cancer types, and that can be expanded to include additional cancer type–specific genes, would be invaluable in research. The Cancer Genome Atlas (TCGA) network performed a systematic analysis research study of more than 3,000 tumors from 12 cancer types to investigate underlying mechanisms of cancer initiation and progression and have identified 127 significantly mutated genes (SMGs) across these tumor types [2].
The xGen Pan-Cancer Hyb Panel is a hybridization capture panel based on the research findings of the TCGA network. The panel comprises xGen Hyb Probes, individually synthesized and quality controlled 120mer oligonucleotides bearing a 5′ biotin modification and manufactured using proprietary Ultramer™ synthesis technology.
Panel composition (target genes)
Table 1. List of genes that are associated with 12 different cancer types organized by gene pathway.
| Cellular process | Gene |
|---|---|
| Transcription factor/regulator | VHL |
| Histone modifier | MLL3 |
| Genome integrity | TP53 |
| RTK signaling | EGFR |
| Cell cycle | CDKNA2 |
| MAPK signaling | KRAS |
| PI(3)K signaling | PIK3CA |
| TGF-β signaling | SMAD4 |
| Wnt/β-catenin signaling | APC |
| Histone | HIST1H1C |
| Proteolysis | FBXW7 |
| Splicing | SF3B1 |
| HIPPO signaling | CDH1 |
| DNA methylation | DNMT3A |
| Metabolism | IDH1 |
| NFE2L | NEF2L2 |
| Protein phosphatase | PPP2R1A |
| Ribosome | RPL22 |
| TOR signaling | MTOR |
| Other | NAV3 |
Product data
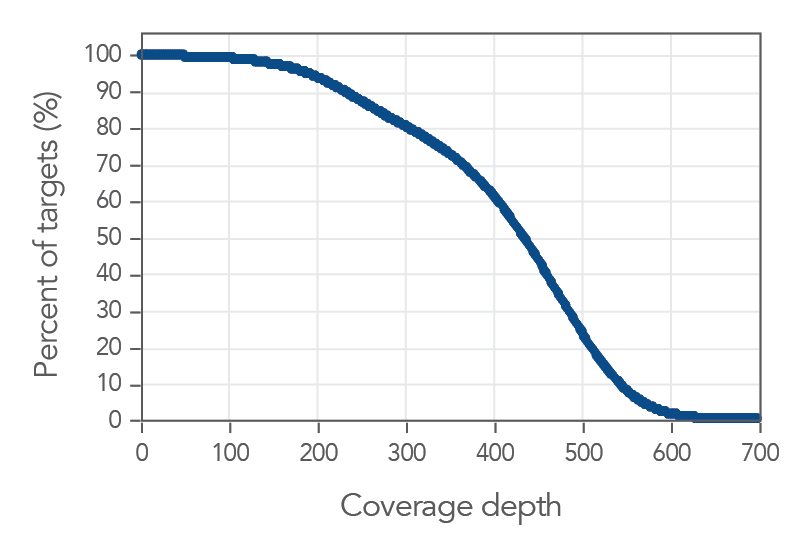
30X coverage depth for 99% of the targets
Figure 1. An example of, deep coverage of targeted regions using xGen Pan-Cancer Hyb Panel. Genomic DNA libraries (n=8) were enriched using the xGen Pan-Cancer Hyb Panel and sequenced on a NextSeq® 550 system using 2 x 150
paired-end reads. Total reads for each sample were sub-sampled to 4M total reads. In all samples, there was >1X coverage for 99.9% of targets and >30X coverage for 99.7% of targets.
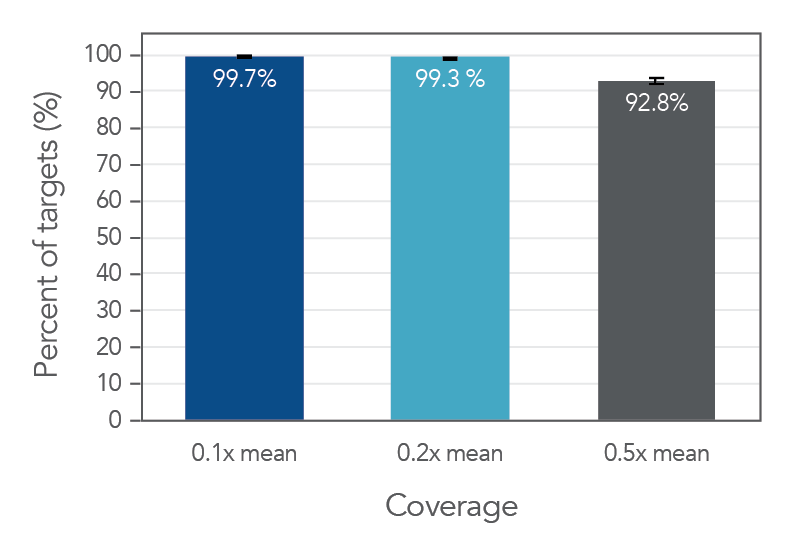
Coverage uniformity for the xGen Pan-Cancer Hyb Panel
Figure 2. An example of high coverage uniformity obtained with xGen Pan-Cancer Hyb Panel. Greater than 0.2X mean coverage is observed for >99.3% of targets. Genomic DNA Illumina TruSeq® HT libraries (n=8) were enriched using the xGen Pan-Cancer Hyb Panel and sequenced on a NextSeq® 550 system using 2 x 150 paired-end reads. Each sample was sub-sampled to 4M total reads.
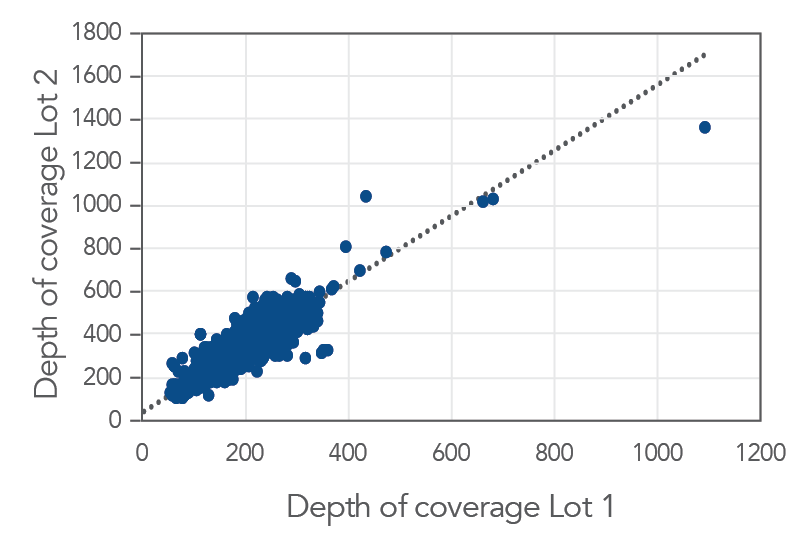
Consistent data from one lot to the next
Figure 3. An example of lot to lot consistency obtained with xGen Pan-Cancer Hyb Panel. Genomic DNA libraries were enriched with two lots of the xGen Pan-Cancer Hyb Panel (n=8 for each lot). The enriched samples were sequenced on the NextSeq 550 sequencing platform using 2 x150 paired-end reads. All samples were sub-sampled to a level of 4M total reads. The probe-by-probe target coverage was averaged for eight replicate samples for each lot of the xGen Pan-Cancer Hyb Panel. A comparison of probe-by-probe target coverage between two lots showed excellent consistency, with an R2 value of 0.8378.
Resources
References
- SEER Cancer Stat Facts: Cancer of Any Site. https://seer.cancer.gov/statfacts/html/all.html. Accessed Sept. 2, 2021.
- Cancer Genome Atlas Research N, Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059-2074.