The CRISPR-Cas9 system has accelerated biomedical research by allowing straightforward and efficient editing of the genome at specific locations. Editing is achieved by generation of double-stranded breaks that either can lead to insertions or deletions and subsequent knockout of genes via the non-homologous end-joining (NHEJ) pathway, or can lead to activation of the homology-directed repair (HDR) process when a template is present. HDR can be used to correct mutations or introduce novel genetic sequences. For instance, HDR could be used to produce fusion proteins.
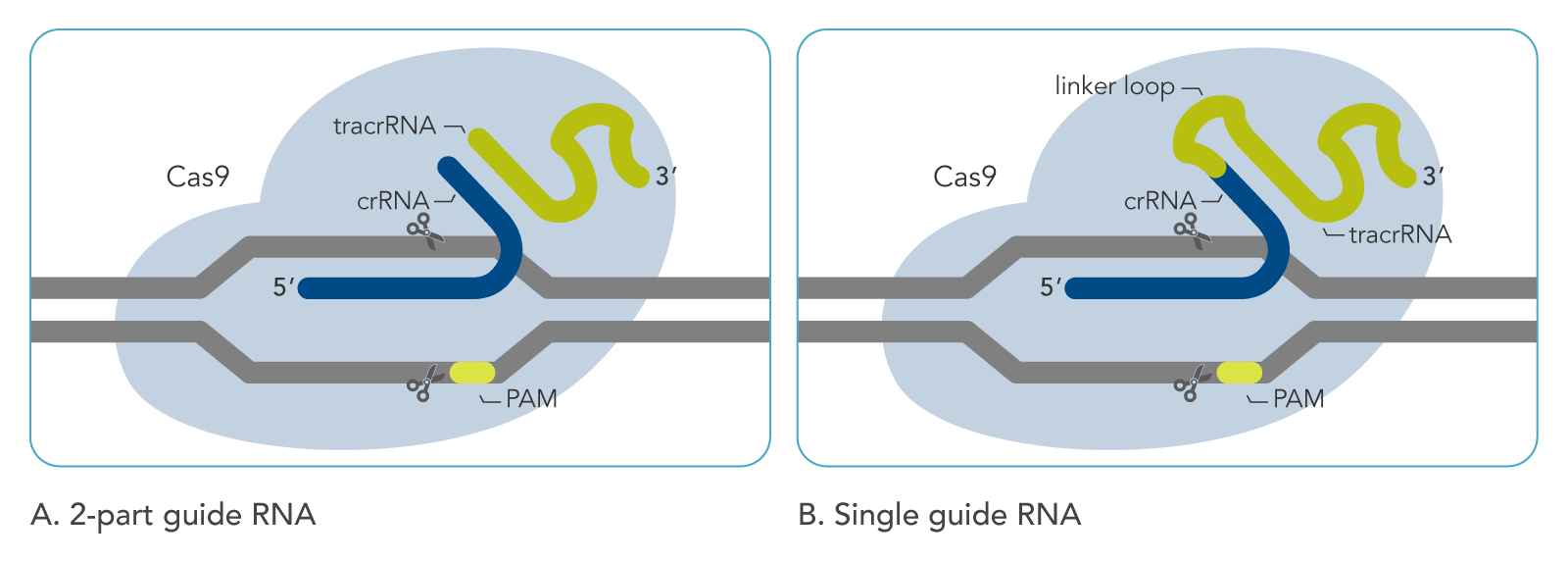
CRISPR-Cas9 editing: crRNA/tracrRNA vs. sgRNA
During CRISPR-Cas9 editing, double-stranded breaks in genomic DNA are generated by the Cas9 protein. The location of DNA cleavage depends on a guide RNA molecule that is necessary for the Cas9 nuclease to be active. This guide RNA consists of a constant region (which binds Cas9) and a variable region, called the spacer, which directs the Cas9 nuclease to a homologous target protospacer location in the genome. In native bacterial immune systems, the Cas9 guide RNA is not just one molecule; it is a duplex made of a tracrRNA and a crRNA (Figure 1A). These two RNA molecules hybridize together by a 16-nt complementary sequence. The constant region is present on the tracrRNA, while the spacer is present on the crRNA molecule.
For use in labs, the tracrRNA and crRNA can be chemically synthesized as RNA oligonucleotides. For Alt-R CRISPR-Cas9 tracrRNAs and crRNAs, synthesis allows for length optimization and introduction of chemical modifications that protect against degradation by endo- and exo-nucleases. Thus, the Alt-R guide RNA complex is stabilized, leading to higher genome editing efficiency. Alternatively, the tracrRNA and crRNA can be synthesized as a single molecule, called a single guide RNA (sgRNA). This molecule includes a loop of a few nucleotides that fuse the crRNA and tracrRNA 16-nt complementary sequences, forming a hairpin-like structure (Figure 1B). As with two-part guide RNAs, chemical modifications can be introduced during synthesis to stabilize Alt-R CRISPR-Cas9 sgRNA.
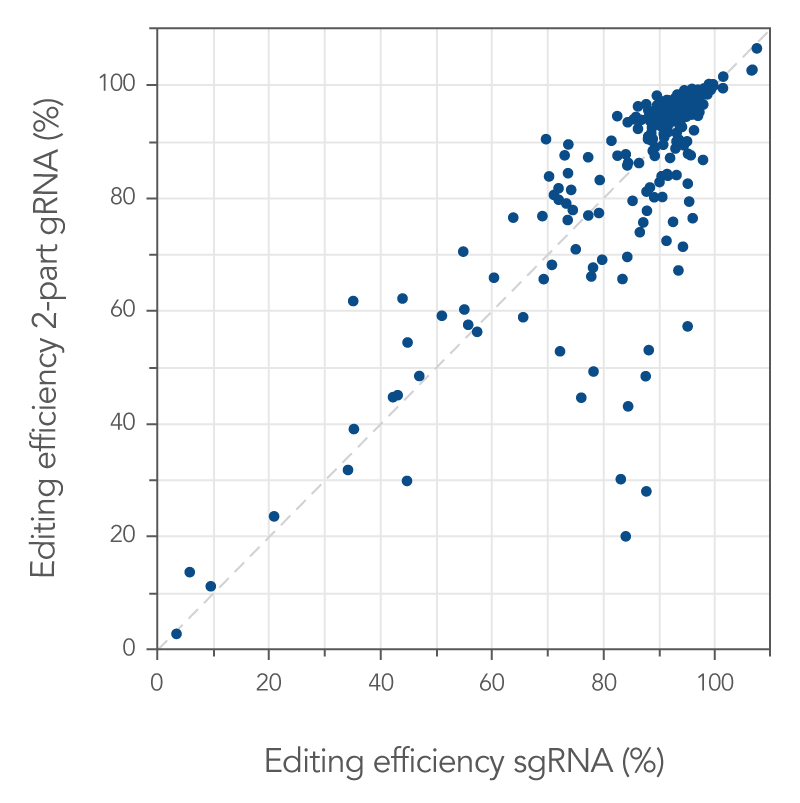
Some reports have stated that single guide RNAs outperform two-part guide RNAs (tracrRNA and a crRNA) in most or all cases. Here, we compare the two types of guide RNA complexes directly. First, 255 target sites were randomly selected across the genome. RNP complexes were made using either two-part or single guide RNA and delivered into Jurkat cells. Genome editing efficiencies were determined. Single guide RNA editing efficiencies for each target site were plotted against the two-part guide RNA editing efficiencies (Figure 2). The majority of target sites (189 out of 255 target sites, 74%) showed genome editing levels of >80%, independent of guide RNA system. These data strongly show the effectiveness of the RNP-based Alt-R CRISPR-Cas9 system.
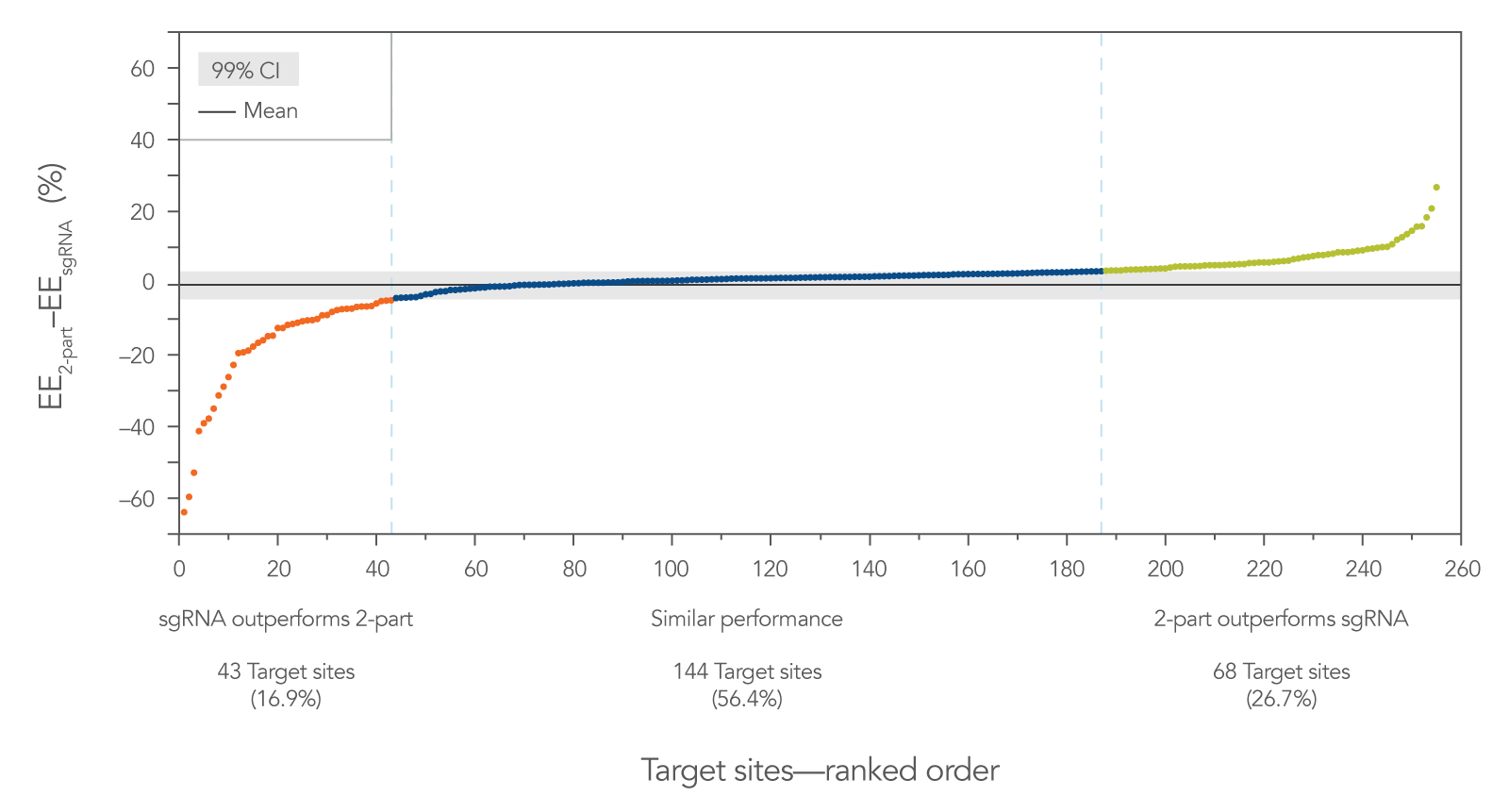
The difference in editing efficiency between two-part guide RNA (tracrRNA and a crRNA) vs. sgRNA was calculated for each target site and plotted in a ranked fashion to visualize the distribution of the differences (Figure 3). For this data set, the 99% confidence interval was calculated (–4.48% ≤ µ2-part – µsgRNA ≤ 3.35%, where µ is the population mean) and is shown as a gray area. The target sites outside of this interval are color-coded. In orange, on the far left of the graph, are the target sites (43 sites, 16.9%) where the sgRNA outperformed the two-part guide RNA. In green, on the far right of the graph, are the target sites (68 sites, 26.7%) where the two-part guide RNA outperformed the sgRNA. Most sites (144 sites, 56.4%) fell within the 99% confidence interval, where both guide RNA formats worked equally well.
These results show that sgRNAs do not always outperform two-part guide RNAs. Rather, this graph shows that the overall editing efficiencies follow a fairly normal distribution. For a number of target sites, sgRNA works better, but for other sites, two-part guide RNA works better.
Considerations when choosing two-part vs. single guide RNAs (sgRNA)
Oligonucleotide synthesis efficiency is negatively correlated to the length of the oligonucleotide. In other words, the longer the oligo, the lower the full-length yield. Consequently, shorter oligos (crRNAs and tracrRNAs) are less expensive than longer oligos (sgRNAs). Conversely, as the two-part system has twice as many ends compared to sgRNAs (4 vs. 2), the two-part system is more susceptible to exonuclease activity. However, this can be addressed by placement of more chemical modifications. It is also worth noting that cell cultures can differ in endogenous nuclease activity. Therefore, we recommend using sgRNA or highly modified two-part guide RNAs (i.e., the Alt-R CRISPR-Cas9 crRNA XT) in environments with high nuclease activity.
The mode of Cas9 delivery also helps determine which guide RNA composition to use. Delivery of Cas9 protein complexed with a guide RNA, known as a ribonucleoprotein complex (RNP), leads to immediate activity in the cell. Therefore, a two-part guide RNA complex can be chosen to form the RNP. When Cas9 protein delivery is indirect via mRNA or plasmid delivery, we recommend using sgRNAs, as they are more stable over time in the intracellular environment. See Table 1 for a summary of these suggestions.
Table 1. Guide RNA recommendations.
|
Situation |
Guide RNA suggestions |
|
Limited budget, no other constraints |
Two-part guide RNA |
|
High endo- or exo-nuclease activity in cells |
Single guide RNA (first choice) OR (next choice if the first choice does not work) two-part guide RNA with many chemical modifications,* OR (third choice, if the above choices do not work) single guide RNA with many chemical modifications** |
|
Delivery of pre-formed Cas9 protein |
Two-part guide RNA, OR single guide RNA (both choices are equally likely to work) |
|
Delivery of mRNA or plasmid coding for Cas9 |
Single guide RNA for longer stability |
|
Low editing efficiency with single guide RNA |
Two-part guide RNA (first choice), OR try another target site (second choice if the first choice does not work) |
|
Low editing efficiency with two-part guide RNA |
Single guide RNA (first choice), OR try another target site (second choice if the first choice does not work) |
* The two-part guide RNA containing many chemical modifications is known as Alt-R CRISPR-Cas9 crRNA XT.
** Single guide RNA with many chemical modifications can be custom-ordered. However, even our standard sgRNAs contain some chemical modifications.
Conclusion
In conclusion, independent of the guide RNA format, high editing levels can be achieved. The data presented here show that for a very small subset of target sites, dramatic differences in editing efficiency can occur.
If you find poor editing efficiency in your experiment, we recommend trying a few different target sites. Alternatively, you can try a different guide RNA format
RUO22-1411_001

