What is CRISPR-Cas9?
CRISPR-Cas9 is a gene editing technology that has a wide range of applications from modifying the genomes of plants to make them more resistant to pathogens [1] to genomic medicine research applications [2-4].
CRISPR stands for Clustered Regularly Interspaced Short Palindromic Repeats, which are a group of short DNA sequences found in the genomes of some archaea and bacteria that function as a defense mechanism [5,6]. When these repetitive segments of DNA are transcribed into RNA called crRNAs, they are complimentary to matching segments of DNA sequences found in invading viruses. The crRNAscan form a complex with a specialized CRISPR nuclease, (e.g., Cas9). This ribonucleoprotein (RNP) complex is able to bind and cleave the target DNA, rendering it inactive [6].
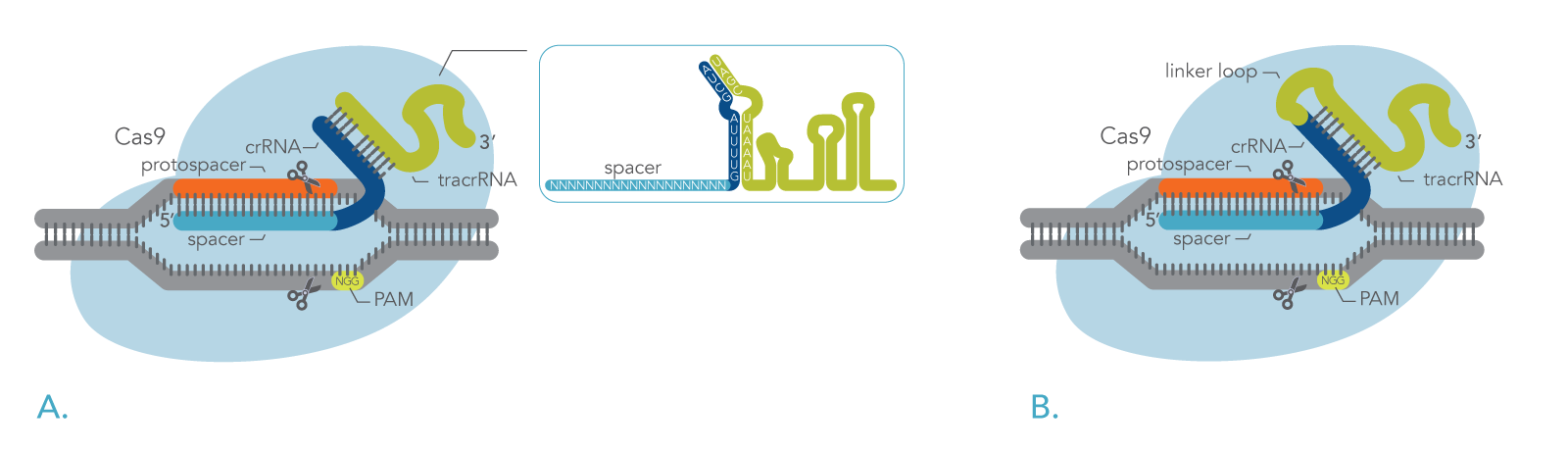
In nature, the Cas9 nuclease binds to a two-part RNA – a crRNA and a trans-activating CRISPR RNA (tracrRNA). Researchers can use modified versions of Cas9 proteins in combination with synthetically generated RNAs to target specific genes, allowing them to better understand the functionality of a gene or to edit genes that may result in a pathological phenotype [3, 7-9]. Figure 1 shows how DNA is cleavage by the CRISPR-Cas9 system using either a two-part guide RNA or a sgRNA.
CRISPR-Cas9 pros and cons
CRISPR-Cas9 is one of many gene editing technologies available to researchers. While it can be an extremely industrious approach for inducing changes to genetic material, it, like any other gene editing application, has several advantages and disadvantages that should be taken into consideration when designing an experiment.
Pros
- Fast, flexible design – Essentially, CRISPR-Cas9 gene editing requires 2 components: the Cas nuclease and a guide RNA. In some cases, researchers opt to use the 2-part crRNA-tracrRNA system while in others they use an sgRNA. Once the guide RNA and endonuclease bind to each other they form a ribonucleoprotein (RNP) complex. It’s within this complex that the guide RNA can bind to the target sequence and editing can occur.
- Multiplexed gene editing – CRISPR-Cas9 can edit multiple genes simultaneously, when the implemented sgRNAs are designed to target different genetic loci [10]. This makes CRISPR-Cas9 gene editing an attractive and efficient approach for manipulating multiple locations in the genome.
- Cost-effective – Because of the reasons listed above, CRISPR-Cas9 can be extremely cost-effective relative to other gene editing approaches such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), which require protein engineering. The simplicity of CRISPR-Cas9 – relying largely on 2 molecules to induce edits – as well as the flexibility available in designing sgRNAs to bind to any/numerous target(s) means that researchers can start editing quickly and make adjustments easily.
Cons
- Delivery limitations – CRISPR-Cas9 has to be successfully delivered into cells in order to induce the desired edits. Like many other gene editing applications, delivery of the Cas9/gRNA complex into a sufficient number of cells can be challenging because all of the components must be delivered into cells at the right concentrations and at the correct point in a cell’s cycle. CRISPR reagent delivery methods include electroporation, lipofection, microinjection, nanoparticles, and viral plasmids.
- Efficiency limitations – Another important limitation to consider when using CRISPR-Cas9 gene editing is that when the Cas9/gRNA complex is taken up by cells, the gene editing activity itself may not occur. This is especially true when the goal is to knock-in or insert material into a gene, a process which relies on homology directed repair (HDR). However, researchers have made significant progress in improving HDR efficiency rates with CRISPR. To read more about improving CRISPR-Cas9 gene editing efficiency, check out the DECODED article, Improving efficiency of homology-directed repair (HDR).
- Off-target effects – Off-target effects occur when the Cas9 nuclease edits an untargeted section of the genome, resulting in unwanted alterations. Off-target effects are a major concern for CRISPR-Cas9 experiments, and they can be challenging to predict. Efforts have been made to improve in silico off-target prediction tools as well as to reduce off-target effects by improving Cas9 nucleases [11,12].
Learn more about CRISPR
CRISPR has proven to be a powerful tool for gene editing and for advancing genomic medicine. CRISPR-Cas9 is one of many options available for gene editing, therefore, it is important to understand the pros and cons of this approach and others when developing an experiment.
You can read more about CRISPR and different gene editing approaches by downloading The CRISPR basics handbook.

