Amplicon sequencing
xGen Amplicon Sequencing Technology has a single-tube workflow to create overlapping super amplicons, which provide coverage of a target space even in the case of primer dropout. This method is applicable to a wide range of research areas and can reduce costs and workflow time. It is also useful for the discovery of rare mutations in complex samples, hard-to-sequence areas, or mutable targets.
xGen™ NGS—made for amplicon sequencing.
Overview
- Integrated library normalization—Normalase™ technology enables a streamlined library balancing and pooling process without the need to quantify samples
- Customizable—add in additional targets to custom amplicon sequencing panels or predesigned panels
- Time saving—single-tube workflow, go from cDNA to normalized library pool in approximately 3 hours
What is amplicon sequencing?
Targeted DNA sequencing methods, such as amplicon sequencing, provide an opportunity for the interrogation of specific regions of the genome so that researchers can obtain information about the genetic regions they care about the most. In amplicon sequencing, highly multiplexed PCR generates amplicons of targeted regions of genomic DNA, after which adapters are added via PCR using indexing primers with either combinatorial dual indexes (CDIs) or unique dual indexes (UDIs). The final library fragments are pooled and sequenced. Index sequences are then used to demultiplex the data bioinformatically, and the sequences of the targeted genomic regions are used for further analysis.
Applications
Amplicon sequencing benefits these areas of research:
- Identification of rare variants
- Identification of hot-spot mutations
- CRISPR genome editing confirmation
- Oncology and virology
- Whole genome sequencing confirmation
- Genotyping
This highly multiplexed PCR approach provides flexibility for a wide range of experimental designs while helping to reduce costs and workflow time. It is also particularly useful for the discovery of rare somatic mutations in complex samples (e.g., tumors mixed with germline DNA) and hard-to-sequence areas (e.g., GC-rich regions).
For an in-depth discussion on this method, see our Amplicon sequencing technical overview.
Predesigned and custom amplicon sequencing panels
IDT offers a wide variety of different predesigned panels, but if research takes you in an uncharted direction, custom amplicon sequencing panels can be created for your specific needs.
Table 1. IDT xGen Amplicon Panels.
| xGen Amplicon Technology | |
|---|---|
| Category | Genetic target† |
| xGen SARS-CoV-2 Amplicon Panels | SARS-CoV-2 genome |
| SARS-CoV-2 S gene | |
| ACE2 gene | |
| xGen Oncology & Inherited Disease Amplicon Panels | 56G Onco v2 |
| 57G PanCancer | |
| BRCA1/2 | |
| BRCA1/2 PALB2 | |
| CFTR | |
| Colorectal genes | |
| EGFR pathway | |
| Lung-associated genes | |
| Lynch Syndrome genes | |
| Myeloid cancer genes | |
| TP53 | |
| xGen Metagenomics Amplicon Panels | 16S rRNA v2 covers V1 through V9 |
| ITS1 | |
| xGen Monkeypox Virus Amplicon Panel | Monkeypox genome |
| Sample ID | Human Sample ID |
| Custom amplicon panel | xGen Custom Amplicon Panel |
| xGen HS Amplicon Technology | |
|---|---|
| Cancer research panel | xGen HS EGFR Pathway Amplicon Panel |
| Amplicon primer sets | |
|---|---|
| Viral research panels | SARS-CoV-2 ARTIC Amplicon Panel |
| SARS-CoV-2 Midnight Amplicon Panel | |
† The 16S Amplicon Panel v2 and xGen ITS1 Amplicon Panel, both of which are covered in this protocol, are separate products that should be purchased individually.
xGen 16S rRNA gene sequencing workflow
Analysis of sequencing data obtained using the xGen 16S Amplicon Panel v2 is presented here as an example of a research question that may be addressed using one of IDT’s predesigned panels. The xGen 16S Amplicon Panel v2 enables library construction from DNA using tiled primer pairs to target the V1–V9 variable regions of the 16S rRNA gene (Figure 1). The xGen 16S Amplicon Panel v2 facilitates the analysis of microbial communities that consist of bacteria and archaea using a single primer pool.
The xGen 16S Amplicon Panel v2, like all the IDT xGen Amplicon Panels (Table 1), utilizes multiple overlapping amplicons in a single tube, using a rapid, two-hour workflow to prepare ready-to-sequence libraries. IDT’s PCR1+PCR2 workflow generates reliable libraries, even from low input quantities (Figure 2). Then libraries can optionally be normalized enzymatically with the xGen Normalase™ technology, which is included with the xGen 16S Amplicon Panel v2. This integrated normalization step can save time and resources.
The xGen Amplicon Panels can also be customized for additional targets by the addition of other predesigned panels or a custom amplicon panel.
The sequencing data generated from xGen Amplicon libraries can be processed and analyzed using the recommendations in Primerclip-A Tool for Trimming Primer Sequences Application Note. For sequences generated with the xGen 16S rRNA v2 Amplicon Panel, the 16S SNAP APP-An automated pipeline for community analysis using multiple 16S rRNA variable regions tool can be used, further streamlining your amplicon based sequencing workflow.
Amplicon workflow
Extraction
Library prep
Choose the appropriate amplicon sequencing panel:
xGen Amplicon Core Kit (found on individual amplicon panel pages)
Sequencing & analysis
Method data
Coverage of xGen Amplicon Panel Technology
To fully analyze complex microbial communities, amplicon sequencing often targets one variable region of the 16S rRNA gene. The IDT xGen Amplicon Panel for 16S rRNA analysis amplifies all nine variable regions of the 16S rRNA gene. To assess the ability of the xGen 16S Amplicon Panel v2 to capture diverse microbial communities, libraries were prepared using a commercially available standard (MSA-1003, ATCC; n = 24) and the xGen 16S Amplicon Panel v2 (covering the V1–V9 regions of the 16S rRNA gene); a panel targeting only the V3–V3 16S rRNA gene regions; and IDT’s legacy Accel-Amplicon 16S+ITS panel. The resulting libraries were sequenced on a MiSeq® (Illumina®) instrument (2 x 150 PE) without the use of the phiX reagent.
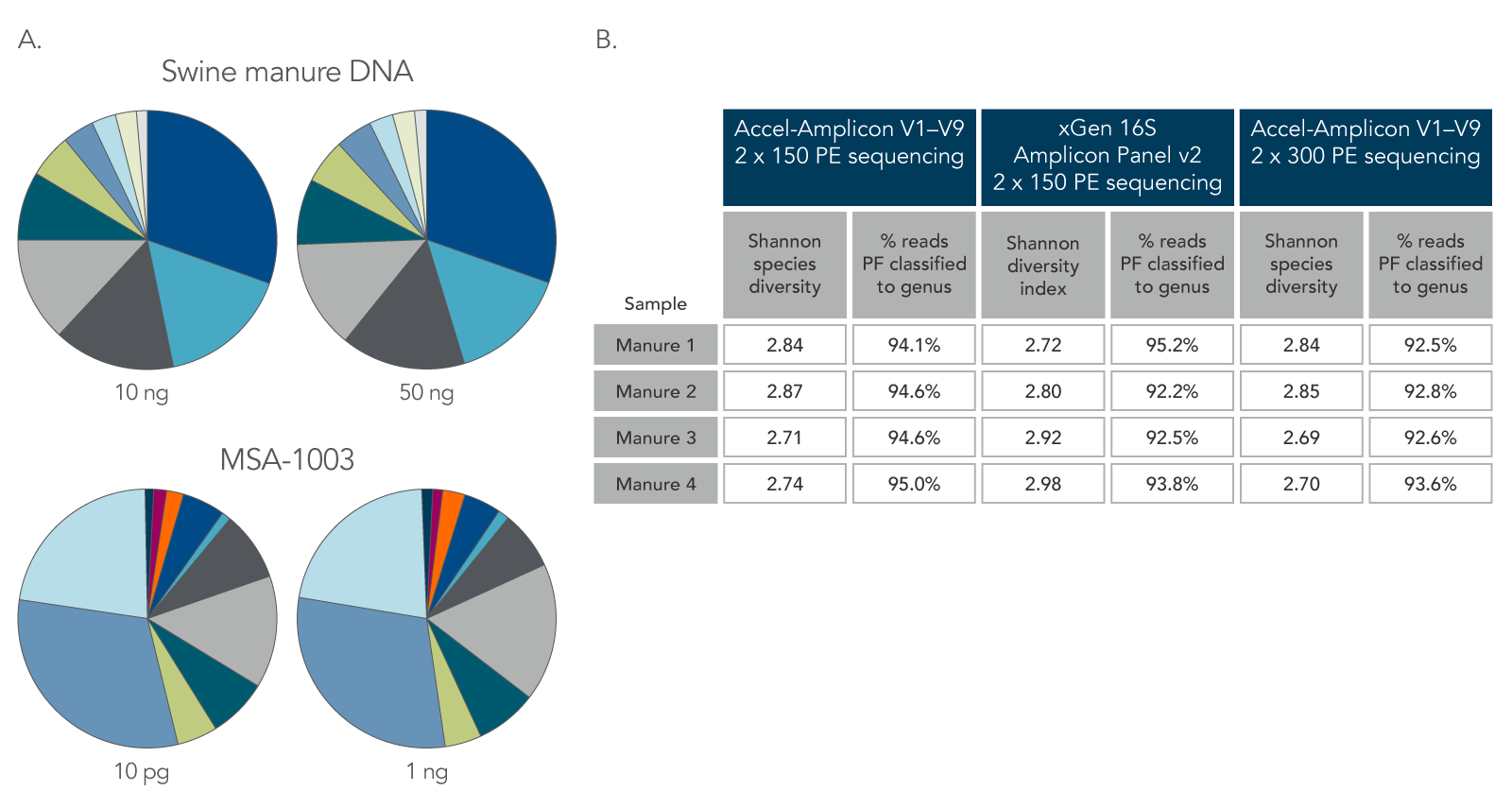
Libraries were also prepared using 10–50 ng of DNA from swine manure (n = 2) and 10 pg and 1 ng of MSA-1003 to establish the flexibility of the sample input into the xGen 16S Amplicon Panel v2. Amplicons were generated using xGen 16S Amplicon Panel v2 protocol and listed cycling conditions, as well as the legacy Accel-Amplicon 16S+ITS panel. Amplicon libraries were sequenced using both 2 x 150 and 2 x 300 PE sequencing chemistries on the MiSeq® platform (Illumina).
The resulting sequences were analyzed using recommendations found in the 16S SNAP APP-An automated pipeline for community analysis using multiple 16S rRNA variable regions.
The resulting sequences from this workflow clearly illustrate the ability of the xGen 16S Amplicon Panel v2 to capture the diversity microbial communities from different sample types, inputs, and using different sequencing chemistries (Figures 1 and 2). This fast, efficient, and flexible workflow provides researchers with all the necessary reagents to produce reliable 16S rRNA gene sequencing datasets across all nine variable regions of the gene.
Figure 1. The xGen 16S Amplicon Panel v2 panel covering V1–V9 regions of 16S rRNA provides an accurate representation of each genus in a commercially available standard (MSA-1003) compared to libraries interrogating the V3–V4 region alone. The legacy Accel-Amplicon 16S+ITS panel and the new xGen 16S Amplicon Panel v2 show similar relative abundance results. Strains were present at levels from 0.02% to 18% in MSA-1003. Presence of the boxed organisms was not observed when only the V3–V4 region was used. Libraries were sequenced on a MiSeq® (Illumina®) instrument without the use of the phiX reagent (n = 1, representative dataset). As shown in the examples in this figure, by not losing reads to phiX or having to sequence deeper due to the use of phased primers, an increased number of samples can be fit on a sequencing run, thereby increasing sequencing efficiency, while still delivering quality data
Figure 2. Consistent function with varying biomass, sample type, and read length. Using the same protocol and cycling conditions, input quantities of 10 ng and 50 ng of DNA from swine manure (A, two top pie charts) resulted in the same relative abundance and types of genera. In addition, the 10 pg DNA sample of a commercially available standard (MSA-1003) had the same relative abundance and types of genera as the 1 ng DNA sample (A, two bottom pie charts). The legacy Accel-Amplicon 16S+ITS panel and the new xGen 16S Amplicon Panel v2 products gave similar results when comparing the 2 x 150 and 2 x 300 PE sequencing read percentages; a comparable number of genera were identified from swine manure samples (B). PF–pass filter.