xGen™ HS EGFR Pathway Amplicon Panel
Targeted sequencing panel for EGFR, BRAF, KRAS, and NRAS
xGen HS EGFR Pathway Amplicon Panel can identify variants with a MAF of ≤1% from ≥ 10 ng of cell-free DNA (cfDNA) or FFPE DNA for Illumina® sequencers.
xGen NGS—made sensitive.
Ordering
- Identifies SNVs and indels down to 0.25% allele frequency
- Compatible with cfDNA and FFPE DNA
- Amplifies from 10–50 ng of cfDNA
- Makes Illumina®-compatible libraries within 3 hours
- Supports research in oncology, graft vs. host disease, and single nucleotide variants in the EGFR pathway
Transform Your NGS Workflow with Automation
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
Request a consultation
Want to learn more about our xGen™ Amplicon Panels—developed with super amplicon technology—and how to customize your panel or spike-in genes in our predesigned panels? Your time is valuable—we’ll prioritize your inquiry and be in touch to discuss it ASAP.
Request a consultationProduct details
The IDT xGen HS EGFR Pathway Amplicon Panel covers contiguous regions of the EGFR gene, and hotspot coverage of BRAF, KRAS, and NRAS with a panel of amplicon-generating primers. The final amplicons average only 136 bp making them compatible with DNA fragments from cell-free DNA (cfDNA) and/or FFPE DNA. The final target size of this panel is 1.5 kbp.
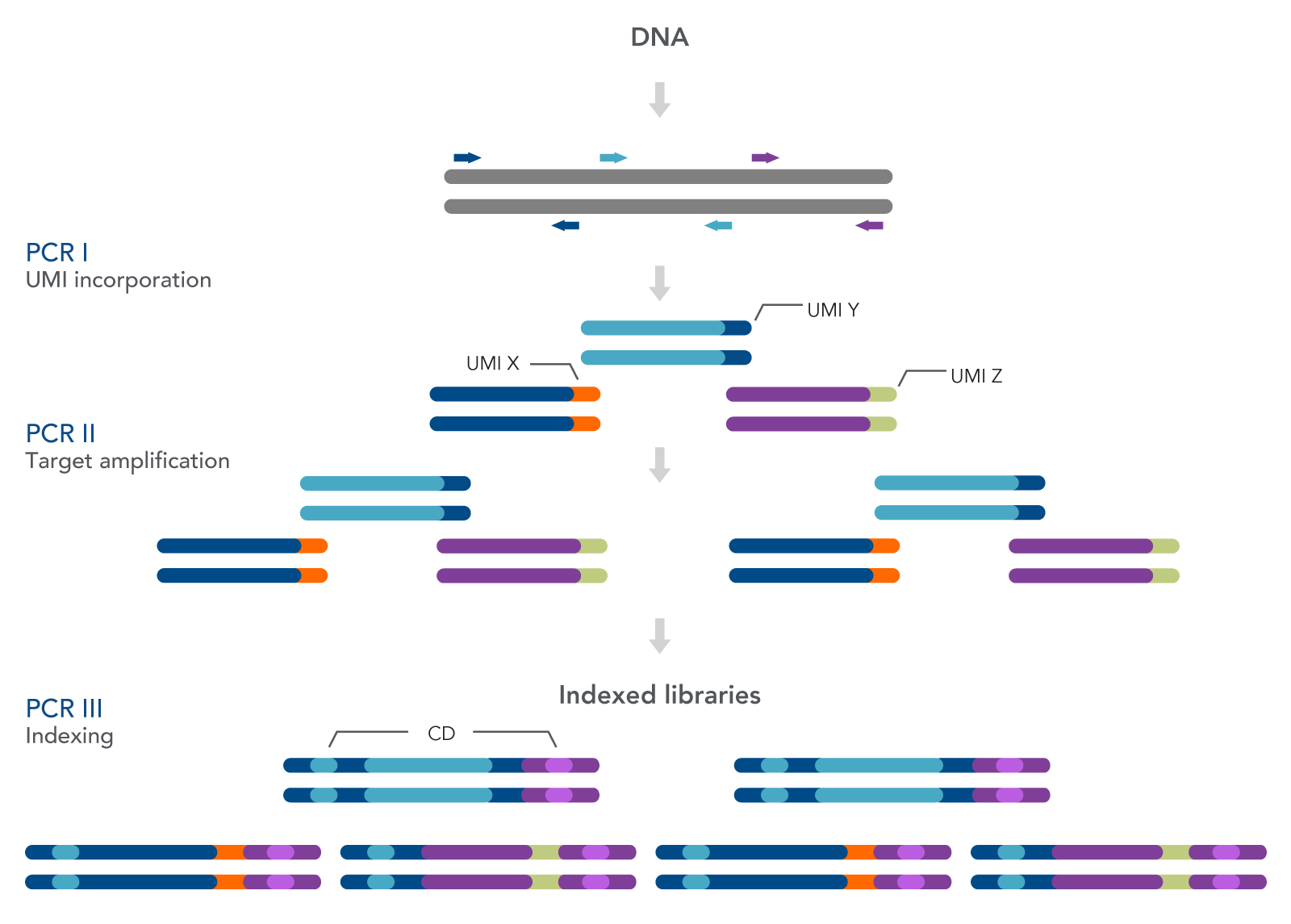
As seen in Figure 1, this workflow includes three PCR steps—one for the incorporation of unique molecular identifiers (UMIs) (PCR I), one for target amplification (PCR II), and one for the addition of combinatorial dual indexed adapters (PCR III), enabling multiplexing of up to 96 unique libraries. Bead-based SPRI® cleanups (Beckman Coulter) are used to purify the sample by removing unused oligonucleotides and changing buffer composition between steps.
Figure 1. The xGen HS EGFR Pathway Amplicon Panels are used to prepare indexed Illumina®-compatible libraries from cfDNA or FFPE DNA. The workflow for preparing the final targeting sequencing libraries includes PCR I, PCR II, and PCR III. The first amplification uses the panel of primers to amplify the targeted genes from the EGFR pathway. The primers also incorporate UMIs. A second amplification step increases the total number of fragments for indexing, while the third PCR completes the indexing.
Product data
To evaluate the xGen HS EGFR Pathway Amplicon Panel, two DNA standards from Seracare were used. These standards contain mutations with known allele frequencies at either 0.5% or 0.25%. Duplicate SeraSeq® ctDNA Reference Material AF (LGC SeraCare) samples at 10 ng were generated with the 0.5% standard and duplicate SeraSeq® ctDNA Reference Material AF (LGC SeraCare) samples at 20 ng were generated with the 0.25% standard, a total of 4 libraries are shown. As seen in Table 1, the observed allele frequency (AF) aligned with the expected allele frequency for the mutations in the reference cfDNA that were targeted by the xGen HS EGFR Pathway Amplicon Panel. Samples were sequenced on a MiniSeqTM (Illumina).
Table 1: Observed vs. expected allele frequency in NGS libraries prepared using the xGen HS EGFR Pathway Amplicon Panel
| 10 ng SeraSeq cfDNA with AF of 0.5% | 20 ng SeraSeq cfDNA with AF of 0.25% | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Mutation | Chr | Position | Expected AF | Observed AF (Rep 1) | Observed AF (Rep 2) | Expected AF | Observed AF (Rep 1) | Observed AF (Rep 2) |
| BRAF | p.V600E | 7 | 140453136 | 0.5% | 0.19% | 0.45% | 0.25% | 0.18% | 0.18% |
| EGFR | p.T790M | 7 | 55249071 | 0.5% | 0.43% | 0.57% | 0.25% | 0.33% | 0.36% |
| EGFR | p.L858R | 7 | 55259515 | 0.5% | 0.86% | 0.43% | 0.25% | 0.26% | 0.25% |
| KRAS | p.G12D | 12 | 25398284 | 0.5% | 0.59% | 0.47% | 0.25% | 0.20% | 0.48% |
| NRAS | p.Q61R | 1 | 115256529 | 0.5% | 0.91% | 0.56% | 0.25% | 0.30% | ND |
| EGFR | p.E746-A750 (delELREA) | 7 | 55242265 | 0.5% | 0.24% | 0.61% | 0.25% | 0.11% | 0.07% |
| EGFR | p.D770-N771 (ins) | 7 | 55249012 | 0.5% | 0.43% | 0.57% | 0.25% | 0.32% | 0.36% |
Resources
Frequently asked questions
Why do my xGen™ Amplicon libraries have low yields and contain primer dimers?
Low library yields and primer dimers may indicate one or more of the following:
- Using too low of an input DNA amount, or using damaged DNA that is poorly amplified
- Not setting up the multiplex PCR master mix and reactions on ice
- Not pre-setting your thermocycler program to temperature before adding samples (letting the thermocycler reach reaction starting temperature with samples in the block)
- Insufficient SPRI clean-ups
How do I analyze xGen™ HS EGFR Pathway Amplicon Panel data?
There are a few key considerations when analyzing sequencing data generated from the xGen HS EGFR Pathway Amplicon Panel with unique molecular identifiers (UMIs).
The first 10 bases in front of Read 2 constitute a UMI. For these first ten bases we recommend trimming them with Trimmomatic (using the CROP option) to make an MID/UMI fastq file for use with the MID pipeline from the fgbio package (Fulcrum Genomics). Before aligning the reads, make sure that the 10 bp UMI (which contains random bases) has been trimmed from 5’ of Read 2.
Also, check that adapter trimming is enabled while setting up the sequencing run. Alternatively, adapter trimming can be performed bioinformatically before analysis.
xGen Custom Amplicon Panels are designed with overlapping amplicons to provide contiguous regions of coverage in a single-tube format. Synthetic primer sequences will be encountered both at the beginning and end of some reads which must be trimmed during analysis. This can be done using a publicly available tool called Primerclip. See our app note Primerclip—A Tool for Trimming Primer Sequences for detailed information.
For more advice, reach out to our Scientific Application Supports team.
Note: A target BED file is provided with purchase of the xGen HS EGFR Pathway Amplicon Panel or the xGen Custom Amplicon Panel.
How long does the xGen™ HS EGFR Pathway Amplicon Panel protocol take to complete?
Can I use my own UDI primers in the xGen™ Amplicon Sequencing workflow?
Yes.
Please contact Scientific Application Support team if you would like assistance confirming compatibility of your own primers with the xGen Amplicon Sequencing workflow.
Note that using your own UDI primers without Normalase™ modifications will make the amplicon libraries incompatible with the downstream Normalase workflow.
Does the xGen™ RNA Library Prep Kit work with damaged or degraded RNA?
Yes.
Please refer to the xGen RNA Library Prep Kit Protocol (Appendix C) for detailed information regarding the changes to RNA fragmentation time and appropriate SPRI bead ratios for samples with RIN scores lower than 7.
We do not recommend generating libraries from starting RNA with a RIN score < 2.Which targets are covered? Which xGen™ Amplicon HS Panels are available?
A single pre-designed panel is available covering the EGFR Pathway and is called the xGen HS EGFR Pathway Amplicon Panel.
See our webpage for target coverage information. View the bioinformatics target BED file and representative data for more information. Or, to design a custom panel, please visit this product page.
What limit of detection (LOD) is achievable with the xGen™ HS EGFR Pathway Amplicon Panel?
The xGen HS EGFR Pathway Amplicon Panel helps to enable variant calling at and below 1% frequency, with allele frequency dependent on the amount of input material available, and the depth of sequencing.
Using 10 ng of input DNA, the xGen HS EGFR Pathway Amplicon Panel consistently identifies variants at 0.5%, with a recommended sequencing depth of at least 60,000x (1M reads), before deduplication with UMIs and 0.25% from 20 ng of input DNA.
Read more on our product page.
Which sample types are compatible with the xGen™ HS EGFR Pathway Amplicon Panel?
The xGen HS EGFR Pathway Amplicon Panel is compatible with (but not limited to) the following sample types:
- Freshly frozen genomic DNA (gDNA)
- Formalin-fixed, paraffin-embedded (FFPE) DNA
- Cell-free DNA (cfDNA)
- High molecular weight DNA
How much DNA is required for the xGen™ HS EGFR Pathway Amplicon Panel?
For the xGen™ HS EGFR Pathway Amplicon Panel, we recommend a minimum of 10 ng input DNA. It is best to quantify input DNA using a qPCR-based assay since it reflects the amplifiable content of the sample.
Note that the input amount affects Limit of Detection (LOD) – 0.5% from 10 ng, and 0.25% from 20 ng.